Abstract
Many human pathogens exploit the actin cytoskeleton during infection, including Toxoplasma gondii, an apicomplexan parasite related to Plasmodium, the agent of malaria. One of the most abundantly expressed proteins of T. gondii is toxofilin, a monomeric actin-binding protein (ABP) involved in invasion. Toxofilin is found in rhoptry and presents an N-terminal signal sequence, consistent with its being secreted during invasion. We report the structure of toxofilin amino acids 69–196 in complex with the host mammalian actin. Toxofilin presents an extended conformation and interacts with an antiparallel actin dimer, in which one of the actins is related by crystal symmetry. Consistent with this observation, analytical ultracentrifugation analysis shows that toxofilin binds two actins in solution. Toxofilin folds into five consecutive helices, which form three relatively independent actin-binding sites. Helices 1 and 2 bind the symmetry-related actin molecule and cover its nucleotide-binding cleft. Helices 3–5 bind the other actin and constitute the primary actin-binding region. Helix 3 interacts in the cleft between subdomains 1 and 3, a common binding site for most ABPs. Helices 4 and 5 wrap around actin subdomain 4, and residue Gln-134 of helix 4 makes a hydrogen-bonding contact with the nucleotide in actin, both of which are unique features among ABPs. Toxofilin dramatically inhibits nucleotide exchange on two actin molecules simultaneously. This effect is linked to the formation of the antiparallel actin dimer because a construct lacking helices 1 and 2 binds only one actin and inhibits nucleotide exchange less potently.
Keywords: actin cytoskeleton, crystal structure, pathogens, analytical ultracentrifugation
The actin cytoskeleton of eukaryotic cells plays an essential role in many processes, including motility and cytokinesis (1). However, certain pathogens, such as Salmonella, Shigella, and Listeria, use the cytoskeleton of host cells as a vehicle during infection (passive invasion) (2–4), whereas others, including Toxoplasma gondii, have evolved their own actin cytoskeletal systems (active invasion) (5–7). It is estimated that approximately one-third of the world's population is infected with T. gondii. This protozoan parasite belongs to the phylum apicomplexa, which includes other human pathogens of major medical importance, such as Plasmodium, the agent of malaria (8, 9). The clinical manifestation of T. gondii is usually benign, but it can be life-threatening for immunocompromised individuals, children, and pregnant women (8).
One of the most abundantly expressed proteins of T. gondii is toxofilin (10). Toxofilin has both actin-monomer sequestering and filament capping activities. Toxofilin is likely to be secreted during invasion because it is apically localized in intracellular tachyzoites (10), is found in rhoptry organelles (11), and presents an N-terminal signal sequence for secretion (Fig. 1A). These findings, together with the observation that overexpression of toxofilin in HeLa cells results in a loss of actin stress fibers (10), suggest that toxofilin (and T. gondii as a result) may disrupt the host actin cytoskeleton during infection. The study of the complex of actin with toxofilin amino acids 69–196 (toxofilin69–196) reported here reveals a number of unexpected features, including stabilization of an antiparallel actin dimer and dramatic inhibition of nucleotide exchange on actin by toxofilin.
Fig. 1.
Structure of the toxofilin69–196–actin complex. (A) Sequence and domain organization of toxofilin. The fragment crystallized with actin is underlined (Gln-69 to Arg-196). The diagram shows the location of helices 1–5, as well as predicted signal sequence, disordered regions, and a C-terminal helix. Helices 1 and 2 interact with a symmetry-related actin (residues in orange background), whereas helices 3–5 form the primary actin-binding region (residues in yellow background). (B) A 1:1 complex in the asymmetric unit of the crystal structure (toxofilin in turquoise and actin in gray). The actin subdomains are numbered 1–4. Some toxofilin residues are shown (red). Close views illustrate the interactions of toxofilin Gln-134 with the nucleotide and helix 3 with the hydrophobic cleft in actin (13). (C) Two symmetry-related complexes interact back-to-back in the structure to form a 2:2 complex. The structure of the WH2 of WASP (15) is superimposed (yellow) to illustrate the resemblance with toxofilin helix 3.
Results
Structure of the Toxofilin69–196–Actin Complex.
Toxofilin is a 245-aa protein (10) (Fig. 1A). The first 27 aa form a signal peptide, which is likely involved in secretion (11). The interaction with actin had been previously mapped to amino acids 69–119 (12). However, for this study the longer fragment 69–196 was selected because sequence analysis suggested that this portion of the molecule consisted of a series of helical segments surrounded by disordered regions at the N and C termini. Crystals of the complex with actin were obtained under different conditions and at two different toxofilin69–196:actin ratios, 1:1 and 1:2 (see Materials and Methods). Crystals of the 1:2 complex (obtained at 20°C) were not pursued in this study because they diffracted the x-rays to low resolution (≈7 Å). The structure reported here to 2.5-Å resolution is that of the 1:1 complex obtained at 4°C (Fig. 1B). Although the asymmetric unit of this crystal form consists of a 1:1 complex, the actual stoichiometry of the complex is 2:2, with two symmetry-related complexes interacting back-to-back in the crystals (Fig. 1C). Interestingly, the 1:1 and 1:2 crystal forms appear to be related, because they share the same symmetry (tetragonal) and two identical unit cell parameters (a and b). In the third direction (c axis), the unit cell of the 1:2 complex is twice as long as that of the 1:1 complex (726 vs. 373 Å). These observations provided the first indication that toxofilin69–196 might interact with two actin molecules in solution (see below).
Most of the actin molecule is defined in the electron density map [supporting information (SI) Fig. 4], with the exception of the DNase I-binding loop (residues 40–51) and the first five and last four residues of the sequence. The last 20 residues of toxofilin69–196 (Arg-177 to Arg-196) are also disordered and do not appear to interact with actin. The conformation of toxofilin69–196 is extended and consists of five helices (Gln-69 to Leu-75, Arg-80 to Gln-92, Thr-103 to Gln-120, Asn-131 to Gln-147, and Ser-149 to Arg-176) connected by loops (Fig. 1B and SI Movie 1). Helices 1 and 2 interact with the symmetry-related actin molecule and partially shield its nucleotide cleft (Fig. 1C). The following 10 aa (93–102) form a loop that connects to helix 3. This loop coincides with the two-fold symmetry axis relating the two interacting complexes.
Helices 3–5 form the primary actin-binding region. Helix 3 binds in the cleft between actin subdomains 1 and 3, a common binding site for multiple, unrelated actin-binding proteins (ABPs) (13). Despite the lack of sequence similarity this interaction is very similar to that of the helix of WASP homology domain 2 (WH2) (Fig. 1C) (14, 15). Like WH2, toxofilin69–196 helix 3 presents hydrophobic residues (Leu-11, Leu-112, and Ile-115) that face the hydrophobic cleft in actin (Fig. 1B, close view). The region C-terminal to helix 3 (residues 121–130) is mostly extended and, like WH2, interacts along the actin surface, climbing toward the pointed end of the actin monomer. However, toxofilin and WH2 follow different paths in this region (Fig. 1C). Toxofilin's path is alongside the interface between the two major actin domains, ending at the nucleotide cleft. The route followed by WH2 coincides with the N-terminal portion of the toxofilin molecule from the symmetry-related complex (albeit with opposite directionalities of the polypeptide chains).
After residue Leu-130, the toxofilin chain takes a sharp (≈90°) turn. From this point on, all of the interactions are with actin subdomain 4. Helices 4 and 5 form an ≈90° elbow (pivot point at Leu-148) and wrap around subdomain 4 (Fig. 1B). Amino acids Ala-137, Val-141, and Leu-145 of helix 4 interact along a cleft on the actin surface lined by residues Leu-216, Tyr-218, Glu-226, Thr-229, Ala-230, Leu-236, and Arg-254. Toxofilin residue Gln-134 in helix 4 penetrates the nucleotide cleft in actin and makes a hydrogen-bonding contact with the NH2 of the ATP (SI Fig. 4 and Fig. 1B, close view).
Toxofilin helix 5 progressively detaches from actin and makes few interactions with it (SI Movie 1). The last interaction observed involves residue Gln-160, which makes a hydrogen-bonding contact with the main chain nitrogen of actin residue Ala-228. The remaining 36 aa of toxofilin69–196 do not interact with actin, and probably as a result the chain is disordered after residue Arg-176. Toxofilin had been predicted to form a coiled coil (10, 12). The current structural and analytical ultracentrifugation (AUC) results (see below) do not support this prediction. It is therefore interesting to note that helix 5 runs antiparallel to itself, i.e., to helix 5 from a symmetry-related complex in the crystal (SI Fig. 5). Although this crystal packing contact resembles an antiparallel coiled-coil dimer, the interaction is too short and lacks the characteristic knobs-into-holes pattern of coiled coils.
Stoichiometry of the Toxofilin69–196–Actin Complex.
The structure suggested that toxofilin might bind two actins in solution (Fig. 1C). AUC was used to test this possibility and to investigate whether toxofilin69–196 forms coiled-coil dimers in solution. AUC runs were carried out in high-ionic-strength, polymerization-compatible F-buffer (see Materials and Methods for buffer composition). Actin and toxofilin69–196 were first run separately and then as mixtures using different ratios (Fig. 2 A and B and SI Fig. 6). The mixing ratios were determined experimentally by using the interference optics of the analytical ultracentrifuge (16). Dilution series of some of the samples were also carried out to detect any self- or heteroassociation effect (Fig. 2B and SI Fig. 6A).
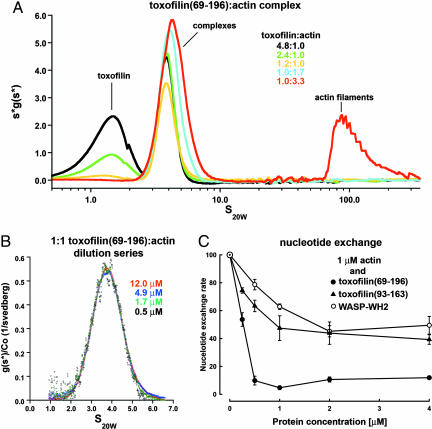
Fig. 2.
Analysis of the toxofilin69–196–actin complex in solution. (A) AUC s*g(s*) plot prepared with the program SEDVIEW (37) of toxofilin69–196:actin mixtures (x axis, sedimentation coefficients on a logarithmic scale in svedbergs at 20°C in water; y axis, sedimentation distribution function multiplied by s*). Each curve represents a combined series of s*g(s*) plots, including all of the scans of a single sedimentation velocity experiment. The area under each curve is proportional to the mass concentration of the sedimenting species. Note that actin filaments appear only in the 1:3.3 mixture (red curve, peak between 65 and 350 S), whereas excess toxofilin69–196 (peaks centered at 1.5 S) appears only in the 4.8:1, 2.4:1, and 1.2:1 mixtures. (B) g(s) plot produced with the program SEDANAL (35) of a dilution series of a 1:1 toxofilin69–196:actin mixture (x axis, sedimentation coefficient; y axis, sedimentation distribution function divided by the loading concentration). The peak corresponding to the 1:1 complex at 3.85 S is stable with dilution and appears alone but presents a minor shoulder at higher S values. This shoulder becomes part of a wider reaction boundary in the 1:1.7 and 1:3.3 mixtures. (C) Inhibition of nucleotide exchange on actin by toxofilin. ε-ATP–actin (1 μM) was mixed with varying concentrations of toxofilin69–196 (●), toxofilin93–163 (▴), or the WH2 of WASP (○), and the fluorescence decay was monitored after addition of 1.1 mM ATP. Note that steady ≈90% inhibition is attained at a toxofilin69–196:actin ratio of 1:2.
Curve fitting of toxofilin69–196 alone at four different concentrations (15, 6, 5, and 2.8 μM) resulted in a single boundary with sedimentation coefficient 1.34 ± 0.01 S and molecular mass 15,184 ± 370 Da (SI Fig. 6 A and B). This value is in excellent agreement with the expected value from sequence (15,043 Da), demonstrating that toxofilin69–196 is a monomer in solution, not a coiled-coil dimer as previously predicted (10, 12). Actin alone formed filaments in F-buffer, characterized by a broad distribution at high S values (60–350 S). An example of such a filament distribution can be observed in a toxofilin69–196:actin mixture containing excess actin (Fig. 2A, red curve).
A 1:1 toxofilin69–196:actin mixture formed a reaction boundary around 3.8 S (Fig. 2B). Although the main peak remained constant with dilution, the data could not be fit to a single 1:1 complex because of the presence of a small concentration-dependent shoulder between 5 and 7 S (detectable only upon fitting). This shoulder suggested the formation of higher-stoichiometry complexes. To investigate this question further, experiments were carried out at different toxofilin69–196:actin ratios (Fig. 2A).
When toxofilin69–196 was added in excess, two overlapping boundaries were observed around 1.5 and 3.8 S (Fig. 2A, black, green, and yellow curves). This experiment fit to a two-species model (SI Fig. 6C), with the first peak corresponding to toxofilin69–196 alone. The calculated mass of the second peak was 55,763 ± 600 Da, corresponding to a 1:1 toxofilin69–196:actin complex (56,961 Da). To assess the relationship between this complex and the crystal structure, the program HYDROPRO (17) was used to calculate a theoretical sedimentation coefficient for the 1:1 complex in the asymmetric unit (Fig. 1B). The calculated S value was 3.73 S, which is in excellent agreement with the observed value (3.85 S), lending support to the formation of a 1:1 complex in solution, with structure similar to that determined here.
When the molar ratio of actin was equal to or higher than that of toxofilin69–196, the boundary around 1.5 S (toxofilin69–196 alone) is no longer observed, indicating that all of the toxofilin69–196 present is bound to actin (Fig. 2 A and B). A new, concentration-dependent reaction boundary formed between 3.8 and 5 S, i.e., shifted to higher S compared with the 1:1 complex. At a 1:1.7 toxofilin69–196:actin ratio (Fig. 2A, cyan curve), no fibers were observed, indicating that toxofilin69–196 sequestered all of the actin present. At a 1:3.3 ratio (Fig. 2A, red curve), 34% of the actin formed filaments at higher S, suggesting that the reaction boundary consisted primarily of a 1:2 complex. Curve fitting was unsuccessful, however, indicating that, in addition to the 1:2 complex, higher-molecular-weight species also form, possibly including the 2:2 complex related by crystal symmetry (Fig. 1C).
In agreement with the structure (Fig. 1C), toxofilin69–196 appears to bind two actins in solution because actin filaments formed only when the molar ratio of actin exceeded two times that of toxofilin69–196 (Fig. 2A, red curve). This result negates a previous report that persisting, although diminished, polymerization occurs at a 2:3 toxofilin:actin ratio (10). To further investigate the stoichiometry of the toxofilin–actin complex, a shorter construct, comprising only the primary actin-binding region (toxofilin93–163), was analyzed. In the presence of a 4-fold molar excess of actin, toxofilin93–163 formed a 1:1 complex with actin (SI Fig. 7), with sedimentation coefficient 3.45 ± 0.01 S and molecular mass 49,600 ± 2,000 Da (expected value 49,990 Da). This result is further evidence that helices 1 and 2 (absent in toxofilin93–163) are necessary, although possibly not sufficient (discussed below), to recruit a second actin to the complex.
Inhibition of Nucleotide Exchange on Actin.
Two observations resulting from the structure are that toxofilin residue Gln-134 interacts with the nucleotide in actin (Fig. 1B) and helix 1 covers the nucleotide cleft of the symmetry-related actin molecule (Fig. 1C). These findings prompted us to study the effect of toxofilin69–196 on nucleotide exchange on actin by monitoring the fluorescence decay upon release of 1,N6-etheno-ATP (ε-ATP) from 1 μM actin in G-buffer. The addition of 0.5 μM toxofilin69–196 resulted in a dramatic ≈90% inhibition of nucleotide exchange on actin (Fig. 2C). As a control, addition of an equivalent amount of a peptide corresponding to the WH2 of WASP resulted in ≈20% inhibition, which is consistent with a previous report (15). Inhibition by the WH2 of WASP stabilized at ≈55% for 2 μM peptide, whereas inhibition by toxofilin69–196 remained constant with concentration at ≈90% (Fig. 2C). It thus appears that toxofilin is a potent inhibitor of nucleotide exchange on actin. In addition, maximum inhibition is attained at a toxofilin69–196:actin ratio of 1:2. This observation is in agreement with the structural and AUC results that suggest that toxofilin binds two actins. Moreover, inhibition appears to be cooperative for these two actins, implying that their antiparallel interaction in the complex potentiates nucleotide exchange inhibition. In support of this proposal, toxofilin93–163, which binds only one actin (SI Fig. 7), inhibited nucleotide exchange to a similar extent as WH2 (Fig. 2C). Analysis of the nucleotide inhibition curves allowed estimating the binding affinities of toxofilin69–196 for the antiparallel actin dimer (≈256 nM) and toxofilin93–163 for the actin monomer (≈207 nM).
Discussion
In the structure of its complex with actin, toxofilin69–196 adopts an extended all-helical fold and interacts with an antiparallel actin dimer related by crystal symmetry (Fig. 3A and SI Movie 1). This interaction might reflect the actual mode of binding in solution, because sedimentation analysis demonstrates the formation of a 1:2 toxofilin69–196:actin complex (Fig. 2A), whereas toxofilin93–163 lacking helices 1 and 2 binds only one actin (SI Fig. 7). Furthermore, toxofilin69–196 produces a dramatic inhibition of nucleotide exchange on actin, and this inhibition also occurs at a 1:2 toxofilin69–196:actin ratio (Fig. 2C). The strong inhibition of nucleotide exchange on actin appears to be potentiated by the formation of the antiparallel actin dimer, which may block domain motions in actin necessary for nucleotide exchange. Indeed, toxofilin93–163, containing a single actin-binding site and comprising residue Gln-134, which interacts with the nucleotide, inhibits nucleotide exchange less potently. Therefore, contrary to our original assumption, direct interaction of Gln-134 with the nucleotide of one actin and steric shielding by helix 1 of the nucleotide cleft of the other actin appear to be less important for nucleotide exchange inhibition.
Fig. 3.
Stabilization of an antiparallel actin dimer by toxofilin and barbed-end filament capping by the toxofilin–actin complex. (A) The structural, AUC, and nucleotide exchange inhibition data are all consistent with the formation of a complex of toxofilin69–196 (turquoise) with an antiparallel actin dimer (gray). The binding interface of the toxofilin molecule on the two actins is colored orange and yellow, according to Fig. 1A. (B) Steric clashes of helices 1, 3, and 5 (indicated by solid arrows) may preclude the binding of toxofilin to the actin filament (20). In contrast, the ternary complex of toxofilin–actin can be added at the barbed end of the filament (but not the pointed end) with very minor clashes involving only helix 5 (indicated by a dashed arrow). Because helix 5 makes few interactions in the complex and appears flexible, it can potentially move, providing a mechanism for filament capping by toxofilin (10).
Given their limited binding interface (Fig. 3A), it appears that toxofilin helices 1 and 2 alone would be insufficient (although necessary) to recruit a second actin unless actin had an intrinsic ability to interact in an antiparallel fashion. Consistent with this proposal, the contact area between actin molecules in the antiparallel dimer is quite substantial, 515 Å2, and includes a symmetric hydrogen-bonding contact between Asp-288 and His-173. On the other hand, the existence of a significant amount of antiparallel actin dimers in solution is well documented (18, 19). Therefore, toxofilin may capitalize on their existence to produce an optimal trap for actin in the unpolymerized state.
Although most of the binding interface of toxofilin69–196 is exposed in Holmes' model of F-actin (20), steric clashes with helix 3, the N terminus of helix 1, and the C terminus of helix 5 may preclude binding to F-actin (Fig. 3B). Interestingly, the structure of toxofilin with the antiparallel actin dimer can be added at the barbed end of the filament (but not the pointed end) with very minor clashes (Fig. 3B). This observation, together with the general flexibility of the toxofilin molecule, which is not organized as a compact domain, may provide a model for filament capping by toxofilin (10).
Toxofilin, like gelsolin (21), vitamin D-binding protein (22), formin (23), WH2 (15), and ciboulot (24), presents a helix (helix 3) that binds in the cleft between actin subdomains 1 and 3 (13). Toxofilin69–196 helix 3 superimposes particularly well with the helix of WH2 (Fig. 1C). Other ABPs also have helices that are predicted to bind in this cleft, including helix 3 of ADF/cofilin (13, 25, 26), the β-tentacle of heterodimeric capping protein (27), and a helix in the p40 (ARPC1) subunit of Arp2/3 complex, thought to dock on an actin subunit of the mother filament during branching (28). What brings so many ABPs to this location? Generally, protein clefts constitute hot spots in molecular recognition (29). Yet, in addition to this common principle of protein–protein interaction, competition of ABPs for a common binding site on actin may be a necessity of regulation, because a large number of ABPs communicate with one another for their respective, and often disparate, signals on actin. However, this cannot explain why a protein from a human pathogen also binds in this cleft. The finding that toxofilin makes part of rhoptry organelles in T. gondii, together with the presence of an N-terminal signal sequence, strongly suggests that toxofilin is secreted during invasion (11). Therefore, toxofilin most likely binds the host cell actin and is unlikely to ever encounter T. gondii's actin, which shares 82% sequence identity with mammalian actin but is not found in rhoptry. Therefore, secretion of toxofilin, which is abundantly expressed, may have the effect of disrupting the host cell cytoskeleton near the site of entry by (i) increasing the pool of unpolymerized actin through its ability to trap antiparallel actin dimers, (ii) capping the barbed end of existing actin filaments, and (iii) interfering with the binding of other ABPs. Whether toxofilin plays such a role during invasion and whether this mechanism is more generally used by other pathogens remain to be demonstrated. The structure reported here can serve as a framework to introduce mutations in toxofilin that would test this hypothesis in vivo.
Other than the interaction of helix 3, the toxofilin–actin structure bears no resemblance to any of the actin complexes studied thus far. In particular, the interaction of helices 4 and 5 that wrap around actin subdomain 4 is unique. Helix 4 fits in a cleft in subdomain 4, which is fully exposed in the filament (Fig. 3B and SI Movie 1). Although less conspicuous than the cleft at the barbed end of the actin monomer (13), the cleft in subdomain 4 may be a preferred site for proteins that bind at the pointed end. A protein whose binding site has been mapped to this cleft by using two-hybrid screening is Aip1 (30). Another obvious candidate to interact in this cleft is tropomodulin.
Phosphorylation of Ser-53 has been reported to lower the affinity of toxofilin for actin (12, 31). Therefore, it is possible that the interaction with actin extends beyond Gln-69 at the N terminus of the toxofilin fragment crystallized here. However, simply assuming that Ser-53 is part of the binding interface cannot explain its role in controlling the actin-binding affinity of toxofilin, because the major binding region consists of helices 3–5. Another possibility is that phosphorylation of Ser-53 triggers a transition toward to a more compact conformation, where the various actin-binding sites are less exposed.
The structural–functional characteristics of toxofilin described here, and the lack of sequence similarity with any known eukaryotic protein, strongly suggest that toxofilin is unique to T. gondii and possibly other apicomplexan parasites. A search for toxofilin homologs in the genomes of three Plasmodium strains (falciparum, yoelii, and chabaudi) reveals hypothetical proteins with ≈27% sequence identity (CAD49159, EAA22346, and CAH75189), localized mainly within the fragment whose structure was determined here. Further research should address whether these proteins are expressed and bind actin.
Materials and Methods
Preparation of Proteins and Peptide.
The cDNAs encoding for toxofilin69–196 and toxofilin93–163 were amplified and cloned between the NdeI and EcoRI sites of the vector pET28a (Novagen, Madison, WI). This plasmid includes an N-terminal polyHis purification tag followed by a thrombin cleavage site. Escherichia coli BL21(DE3) cells (Invitrogen, Carlsbad, CA) were transformed with the toxofilin constructs and grown in LB media at 37°C until the OD at 600 nm reached a value of 0.8. Expression was induced by addition of 1 mM isopropylthio-β-d-galactoside and carried out overnight at 20°C. The proteins were first purified on an affinity Ni-NTA agarose resin (Qiagen, Valencia, CA), followed by dialysis against 20 mM Tris·Cl (pH 6.8), 50 mM NaCl, and 1 mM DTT and purification on a MonoS column (Amersham Biosciences, Piscataway, NJ). His-tag removal was carried out by digestion with thrombin at room temperature in 20 mM Tris·Cl (pH 7.5), 300 mM NaCl, and 1 mM MgCl2. Thrombin and the cleaved His-tag peptides were removed on a benzamidine column (Amersham Biosciences), followed by purification through a Ni-NTA agarose resin (Qiagen). Actin was prepared and labeled with pyrene as described (32).
Crystallization, Data Collection, and Structure Determination.
Toxofilin69–196 and actin in G-buffer (2 mM Tris·Cl, pH 7.5/0.2 mM CaCl2/0.2 mM ATP) were mixed at 1:1 and 1:2 molar ratios, followed by dialysis in 20 mM Tris·Cl (pH 7.5), 200 mM NaCl, 0.2 mM CaCl2, 0.2 mM ATP, and 5 mM DTT. The complexes were then concentrated to ≈10 mg·ml−1 by using a Centricon device (Millipore, Billerica, MA) and centrifuged at high speed (100,000 × g) before crystallization. Crystals of the 1:1 and 1:2 toxofilin69–196–actin complexes were obtained by using the vapor diffusion method under two different sets of conditions. Crystals of the 1:2 complex were obtained at 20°C by mixing 2 μl of protein solution and 2 μl of well solution containing 1 M (NH4)2SO4, 100 mM Hepes (pH 7.0), and 0.5% polyethylene glycol 8000. These crystals had extremely large unit cell parameters (≈726 Å) and diffracted the x-rays to low resolution (≈7 Å). Additional search for conditions resulted in crystals of the 1:1 complex at 4°C from a well solution containing 8% polyethylene glycol 4000, 10% glycerol, and 100 mM sodium acetate (pH 4.6). A complete x-ray data set was collected to 2.30-Å resolution at beamline A1 of the Cornell High Energy Synchrotron Source (Ithaca, NY). The quality of the diffraction data were deemed appropriate only to 2.5-Å resolution (Rmerge < 30%). The diffraction data were indexed and scaled with the program HKL2000 (HKL Research, Charlottesville, VA) (Table 1). A molecular replacement solution for the actin portion of the structure was obtained with the program AMoRe (33), using as search model the 2.0-Å resolution structure of actin complexed with the WH2 of WASP (15). Model building and refinement were carried out with the program COOT (34) and the CCP4 program REFMAC (Table 1). The refinement converged to R-factor and Rfree values of 22.9 and 28.4%. These values are slightly higher than what could be expected at 2.5-Å resolution, which may be because of general disorder in the structure. Indeed, 41 aa were not observed in the electron density map, and the overall temperature factor of the structure is 52 Å2.
Table 1.
Crystallographic data and refinement statistics
| Diffraction statistics | |
|---|---|
| Space group | P 41 2 2 |
| Cell parameters | |
| a, b, c, Å) | 54.523, 54.523, 363.100 |
| α, β, γ, ° | 90.0, 90.0, 90.0 |
| Data resolution, Å | 50.0–2.5 (2.54–2.50) |
| Completeness, % | 100.0 (100.0) |
| Redundancy | 34.3 (27.6) |
| Rmerge,* % | 9.1 (26.8) |
| Average I/σ | 38.6 (14.0) |
| Refinement statistics | |
| Refinement resolution, Å | 50.0–2.5 (2.565–2.50) |
| R-factor,† % | 22.9 (26.0) |
| Rfree‡, % | 28.4 (33.0) |
| rmsd | |
| Bond length, Å | 0.010 |
| Bond angles, ° | 1.305 |
| Average B-factors, Å2 | |
| All/actin/toxofilin/solvent | 51.9/48.1/64.6/48.7 |
| Protein Data Bank ID code | 2Q97 |
Values in parentheses correspond to highest-resolution shell.
*Rmerge = Σ(I − 〈I〉)/ΣI. I and 〈I〉 are the intensity and the mean value of all the measurements of an individual reflection
†R-factor = Σ|Fo − Fc|/Σ|Fo|. Fo and Fc are the observed and calculated structure factors.
‡Rfree, R-factor calculated for a randomly selected subset of the reflections (5%) that were omitted during the refinement.
AUC Analysis.
Sedimentation velocity experiments were conducted in an Optima XL-I ultracentrifuge (Beckman, Palo Alto, CA), using either an An60 Ti four-hole rotor or an An50 Ti eight-hole rotor. Data were acquired with the interference optics system using sapphire windows. To improve the signal-to-noise ratio, the cell assembly was enhanced by using double-sector, meniscus-matching, 12-mm, aluminum-filled Epon centerpieces and interference slit window holders on the top window (Biomolecular Interaction Technologies Center, Durham, NH). Toxofilin69–196–actin mixtures were prepared at different molar ratios. The mixing ratios, which were initially estimated by using theoretically calculated extension coefficients, were subsequently determined more precisely (reported values) by using the interference optics of the analytical ultracentrifuge (16). Toxofilin69–196, toxofilin93–163, and actin samples were analyzed alone and as mixtures. The samples were subjected to overnight dialysis in F-buffer (20 mM Tris·Cl, pH 7.5/100 mM NaCl/0.2 mM ATP/0.2 mM CaCl2) and then spun at 6,000 rpm for 5 min to match the menisci. After this step, the rotor was removed, shaken, and placed back in the chamber to allow for the temperature to equilibrate for at least 1 h. Velocity experiments were conducted at 50,000 rpm and 20°C. Sedimentation velocity data were analyzed by using the program SEDANAL (35). A curve was deemed acceptable only if the standard deviation was less than or equal to the previously measured optical noise of the system (6 × 10−3 fringes). The program SEDNTERP (36) was used to estimate the hydration, size, and asymmetry of the specimens and to covert the sedimentation coefficients to the reported S20W values (S value at 20°C in water).
Nucleotide Exchange Assay.
The rate of nucleotide exchange on actin was measured by monitoring the fluorescence decay resulting from the release of 1,N6-etheno-ATP (ε-ATP; MP Biomedicals, Irvine, CA) from G-actin at room temperature. ε-ATP-labeled G-actin was prepared by extensive dialysis in G-buffer containing ε-ATP (instead of ATP). Aliquots of ε-ATP–G-actin were incubated with various concentrations of toxofilin69–196, toxofilin93–163, or the WH2 of WASP, and the substitution reaction was started by adding 1.1 mM ATP in G-buffer. The reaction was monitored at 412 nm with excitation at 360 nm by using a Cary Eclipse Fluorescence Spectrophotometer (Varian, Palo Alto, CA). The nucleotide exchange rate was calculated from the initial slope of the fluorescence decay and expressed as a fraction of the exchange rate in the absence of added toxofilin69–196.
Supplementary Material
Acknowledgments
We thank Francois Ferron for help with the preparation of SI Movie 1. This work was supported by National Institutes of Health Grants GM073791 and HL075404. Use of the Cornell High Energy Synchrotron Source and Macromolecular Crystallography Facility was supported by National Science Foundation Grant DMR-0225180 and National Institutes of Health Grant RR-01646.
Abbreviations
- ABP
actin-binding protein
- WH2
WASP homology domain 2
- AUC
analytical ultracentrifugation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2Q97).
This article contains supporting information online at www.pnas.org/cgi/content/full/0705794104/DC1.
References
- 1.Pollard TD, Borisy GG. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 2.Patel JC, Galan JE. Curr Opin Microbiol. 2005;8:10–15. doi: 10.1016/j.mib.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Galan JE, Zhou D. Proc Natl Acad Sci USA. 2000;97:8754–8761. doi: 10.1073/pnas.97.16.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gouin E, Welch MD, Cossart P. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Morrissette NS, Sibley LD. Microbiol Mol Biol Rev. 2002;66:21–38. doi: 10.1128/MMBR.66.1.21-38.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morisaki JH, Heuser JE, Sibley LD. J Cell Sci. 1995;108:2457–2464. doi: 10.1242/jcs.108.6.2457. [DOI] [PubMed] [Google Scholar]
- 7.Dobrowolski JM, Sibley LD. Cell. 1996;84:933–939. doi: 10.1016/s0092-8674(00)81071-5. [DOI] [PubMed] [Google Scholar]
- 8.Kim K, Weiss LM. Int J Parasitol. 2004;34:423–432. doi: 10.1016/j.ijpara.2003.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubremetz JF. Trends Microbiol. 1998;6:27–30. doi: 10.1016/S0966-842X(97)01165-7. [DOI] [PubMed] [Google Scholar]
- 10.Poupel O, Boleti H, Axisa S, Couture-Tosi E, Tardieux I. Mol Biol Cell. 2000;11:355–368. doi: 10.1091/mbc.11.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, Dunn JD, Ferguson DJ, Sanderson SJ, Wastling JM, Boothroyd JC. J Biol Chem. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- 12.Jan G, Delorme V, David V, Revenu C, Rebollo A, Cayla X, Tardieux I. Biochem J. 2007;401:711–719. doi: 10.1042/BJ20061324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez R. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Lee SH, Kerff F, Chereau D, Ferron F, Klug A, Dominguez R. Structure (London) 2007;15:145–155. doi: 10.1016/j.str.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Proc Natl Acad Sci USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babul J, Stellwagen E. Anal Biochem. 1969;28:216–221. doi: 10.1016/0003-2697(69)90172-9. [DOI] [PubMed] [Google Scholar]
- 17.Garcia De La Torre J, Huertas ML, Carrasco B. Biophys J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millonig R, Salvo H, Aebi U. J Cell Biol. 1988;106:785–796. doi: 10.1083/jcb.106.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bubb MR, Govindasamy L, Yarmola EG, Vorobiev SM, Almo SC, Somasundaram T, Chapman MS, Agbandje-McKenna M, McKenna R. J Biol Chem. 2002;277:20999–21006. doi: 10.1074/jbc.M201371200. [DOI] [PubMed] [Google Scholar]
- 20.Holmes KC, Popp D, Gebhard W, Kabsch W. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 21.Irobi E, Burtnick LD, Urosev D, Narayan K, Robinson RC. FEBS Lett. 2003;552:86–90. doi: 10.1016/s0014-5793(03)00934-7. [DOI] [PubMed] [Google Scholar]
- 22.Otterbein LR, Cosio C, Graceffa P, Dominguez R. Proc Natl Acad Sci USA. 2002;99:8003–8008. doi: 10.1073/pnas.122126299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 24.Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Preat T, Knossow M, Guittet E, Carlier MF. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- 25.Wriggers W, Tang JX, Azuma T, Marks PW, Janmey PA. J Mol Biol. 1998;282:921–932. doi: 10.1006/jmbi.1998.2048. [DOI] [PubMed] [Google Scholar]
- 26.Mannherz HG, Ballweber E, Galla M, Villard S, Granier C, Steegborn C, Schmidtmann A, Jaquet K, Pope B, Weeds AG. J Mol Biol. 2007;366:745–755. doi: 10.1016/j.jmb.2006.11.100. [DOI] [PubMed] [Google Scholar]
- 27.Narita A, Takeda S, Yamashita A, Maeda Y. EMBO J. 2006;25:5626–5633. doi: 10.1038/sj.emboj.7601395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski RA, Luscombe NM, Swindells MB, Thornton JM. Protein Sci. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. J Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delorme V, Cayla X, Faure G, Garcia A, Tardieux I. Mol Biol Cell. 2003;14:1900–1912. doi: 10.1091/mbc.E02-08-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper JA, Walker SB, Pollard TD. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 33.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 34.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Stafford WF, Sherwood PJ. Biophys Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 36.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. In: Analytical Ultracentrifugation in Biochemistry and Polymer Sciences. Harding SE, Rowe AJ, Horton JC, editors. Cambridge, UK: R Soc Chem; 1992. pp. 90–125. [Google Scholar]
- 37.Stafford WF, Braswell EH. Biophys Chem. 2004;108:273–279. doi: 10.1016/j.bpc.2003.10.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.