Abstract
The bacterial outer membrane protein G (OmpG), a monomeric pH-gated porin, was overexpressed in Escherichia coli and refolded in β-octyl glucoside micelles. After transfer into dodecylphosphocholine micelles, the solution structure of OmpG was determined by solution NMR spectroscopy at pH 6.3. Complete backbone assignments were obtained for 234 of 280 residues based on CA, CB, and CO connection pathways determined from a series of TROSY-based 3D experiments at 800 MHz. The global fold of the 14-stranded β-barrel was determined based on 133 long-range NOEs observed between neighboring strands and local chemical shift and NOE information. The structure of the barrel is very similar to previous crystal structures, but the loops of the solution structure are quite flexible.
Keywords: porin, membrane protein folding, dodecylphosphocholine micelle
Membrane proteins (MP) continue to be prime drug targets because they function as receptors for cellular communication and gatekeepers for ion and substrate transport across cell membranes. Despite the tremendous progress that has been made in the past decade in solving an increasing number of MP structures (1), the total number of structure determinations of MPs still lags far behind the corresponding number for soluble proteins. Most MP structures solved to date have been and probably will continue to be determined by x-ray crystallography. However, some MPs resist crystallization, and some functional aspects of MP structure as well as their dynamics are better studied in solution. Solution NMR spectroscopy is the ideal tool to study the structural dynamics of proteins and recently has been successfully applied to such studies of very large protein complexes (2). Recent advances in instrumentation and pulse program implementation have proven solution NMR to also be a valuable method to determine the structures and dynamic aspects of polytopic integral MPs (3–9). To study MPs by solution NMR, they are usually reconstituted into micelles formed by nondenaturing detergents that are thought to mimic the natural membrane environment quite well, while retaining the overall size of the protein/micelle complex still manageable by solution NMR techniques (10). Various detergents and detergent/lipid combinations have been reported in solution NMR structural studies of MPs with variable degrees of success (11–13). Despite many recent efforts and successes in this area, it is clear that MP sample preparation remains one of the most critical steps in obtaining spectra of a quality that is suitable for a structure determination. In general, α-helical MPs have to be natively expressed and extracted from the cell membranes, whereas β-barrel MPs can often be produced in insoluble inclusion bodies and then refolded in micellar systems that are suitable for solution NMR (3, 4, 6).
The outer membrane protein G (OmpG) is an integral MP that resides in the outer membrane of Gram-negative bacteria. It functions as a monomeric porin and facilitates the uptake of large oligosaccharides (14). Because the primary facilitator of oligosaccharide uptake in Escherichia coli is LamB, OmpG is not normally expressed in LamB-expressing cells. However, when LamB is defective or down-regulated, OmpG is up-regulated (15). When reconstituted in planar lipid bilayers, OmpG forms monomeric channels with a single channel conductivity of 0.8 nS in 1 M NaCl (16). The crystal structure of OmpG was determined very recently by two groups (17, 18). As predicted (16), OmpG forms a 14-stranded β-barrel with seven large loops extending into the extracellular space. One of these studies shows that OmpG adopts an open state at neutral pH (pH 7.5) and a closed state at acidic pH (pH 5.6). The conversion from the open to the closed state involves the folding of loop 6 into the barrel lumen and is associated with an unzipping of hydrogen bonds between strands 12 and 13 triggered by the protonation of two adjacent histidines at the lower pH.
Our interest in OmpG is threefold. It serves as an excellent model for structural studies by solution NMR of a MP of unprecedented large size (33 kDa, 280 aa residues) compared with MP structures that have been previously studied by solution NMR. Like OmpA (19), OmpG also serves as an excellent model for studying the folding of a large β-barrel MP into lipid bilayers (20). Finally, and importantly, OmpG as a monomeric porin is an attractive candidate for its use as an engineerable pore for the design of biosensors.
Results and Discussion
OmpG Refolding and NMR Sample Preparation.
Because OmpG was purified in an unfolded form, we first had to find optimal conditions for refolding the protein with high efficiency and stability for extended NMR experiments at elevated temperatures. This challenge turned out to be more difficult than anticipated from experience with smaller β-barrels such as OmpA (21). We tried the short-chain phospholipid 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) and Fos-cholines with different acyl chain lengths from 8 to 14, which also include dodecylphosphocholine (DPC). We also tried two alkyl-glycosides, namely β-OG and n-dodecyl-β-d-maltopyranoside (DDM), at various concentrations, pH values, and temperatures. Folding was examined by a shift of the apparent molecular mass from 33 to 28 kDa in samples that were not boiled before loading on SDS gels (20). In many cases, folding was either not efficient or the samples were not stable at higher temperatures. In addition, folding aids, such as urea, arginine, ethanol, glucose, and glycerol, were tried at various concentrations. Ultimately, we achieved best refolding when a concentrated OmpG sample in 8 M urea was added dropwise to a 70 mM solution of β-OG micelles at pH 9 and room temperature, followed by incubation at 37°C for 2 more days. An SDS gel of a typical refolding experiment in β-OG is shown in Fig. 1a. The efficiency of refolding was better than 95% based on gel densitometry.
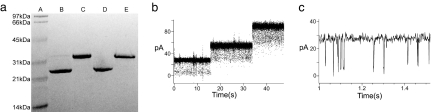
Fig. 1.
(a) SDS/PAGE gel-shift assay showing the refolding efficiency of OmpG in β-OG. Lane A, molecular mass standards; lane B, refolded OmpG in β-OG; C, boiled refolded OmpG; D, refolded OmpG treated with proteinase K; E, denatured OmpG in 8 M urea. (b) Planar lipid bilayer recordings of OmpG in diPhPC bilayer. (c) The open pores fluctuate with briefly closed states as seen on an expanded time scale.
The pore function of refolded OmpG was tested by electrophysiological recordings in a planar lipid bilayer recording setup. In these experiments, the refolded OmpG in β-OG was added to a preformed planar lipid bilayer between two chambers, and the electrical current was measured after voltage was applied across the lipid bilayer. A pore activity that is typical of porins was observed almost immediately after applying the voltage (Fig. 1b). Individual molecules of OmpG open and remain open at different times, giving rise to a stepwise current–time profile. The individual steps correspond to a conductance value of 0.7 nS for a single OmpG molecule, which is close to the 0.8 nS that have been measured before for OmpG under slightly different conditions (16). This behavior is very different from the channel activity of OmpA, which fluctuates between open and closed forms with much lower open probabilities and mean conductance values of ≈30 pS under similar conditions (22). Once open, the OmpG channel also fluctuates but with only very brief closing times (Fig. 1c).
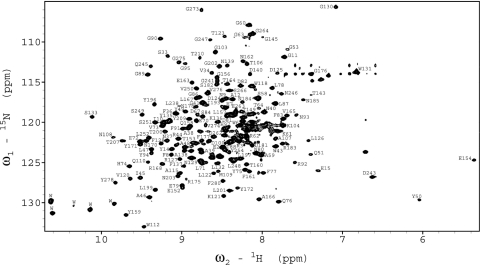
Although initial NMR experiments of refolded OmpG/β-OG samples were quite promising, showing ≈200 resolved cross-peaks spread between 9.8 and 7.5 ppm in the proton dimension of the 15N-1H TROSY spectrum, samples in β-OG ultimately turned out not to be stable enough for long-term triple-resonance experiments at 40°C. Therefore, refolded OmpG/β-OG samples were exchanged into DPC micelles, which was selected because its phosphocholine headgroup matches that of the most abundant phospholipid in the lipid bilayer of biological membranes and because its acyl chain is long enough to cover the hydrophobic surface area of outer MPs in a similar fashion as phospholipids would in a lipid bilayer. DPC also has been used successfully in solution NMR studies of other MPs (12, 13). The OmpG/DPC samples showed an ≈10% increase of the average peak linewidth in the TROSY spectrum compared with OmpG/β-OG samples. However, importantly, spectra in both detergents showed similar chemical shifts for most peaks. The OmpG/DPC samples turned out to be very stable: the average peak linewidth changed very little after weeks of measurements at 40°C. The tradeoff of the increased sample stability against the increased linewidth ultimately proved worthwhile because the spectra were still well enough resolved to permit the assignment of close to 90% of all residues. A typical 15N-1H TROSY spectrum recorded at 800 MHz is shown in Fig. 2.
Fig. 2.
15N-1H TROSY–HSQC spectrum of OmpG in DPC micelles at pH 6.3, collected at 800 MHz and 40°C. Assignments of backbone amides are denoted by one-letter amino acid abbreviations followed by their sequence numbers.
NMR Assignments and Measurements of NOEs.
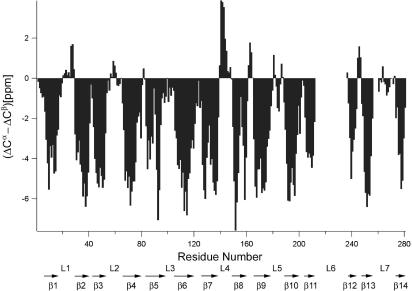
Based on the CA, CB, and CO connection pathways provided by six TROSY-type through-bond triple resonance experiments, 234 (including 8 prolines) of the 280 residues of OmpG (excluding the leading methionine) were completely assigned, 9 residues were partially assigned, and another 37 residues were unassigned. The Cα and Cβ secondary chemical shifts indicated the existence of multiple long β-strands. Fig. 3 shows three-residue averaged (ΔCα–ΔCβ) secondary chemical shifts plotted as a function of amino acid sequence numbers, where (ΔCα–ΔCβ)i were calculated as 1/3(ΔCi−1α + ΔCiα + ΔCi+1α − ΔCi−1β − ΔCiβ − ΔCi+1β) (23). The pattern of this figure clearly reveals the existence of 14 β-strands as indicated by arrows on the bottom. Based on large positive (ΔCα–ΔCβ) values between β7 and β8, a single turn of α-helix might exist in this region. For structure calculations, φ and ψ dihedral angles were predicted from the calibrated chemical shifts by using the TALOS program (http://spin.niddk.nih.gov/NMRPipe/talos). A total of 131 residues gave good fits and yielded 262 dihedral angles.
Fig. 3.
Three-bond averaged secondary chemical shifts, where (ΔCα − ΔCβ)i = 1/3(ΔCi−1α + ΔCiα + ΔCi+1α − ΔCi−1β − ΔCiβ − ΔCi+1β), versus residue sequence numbers i. Each individual ΔC value on the right-hand side of the equation is the chemical-shift deviation from its respective random coil value. Putative β-strands indicated by characteristic negative (ΔCα − ΔCβ) values are denoted by arrows with β-strand numbers underneath.
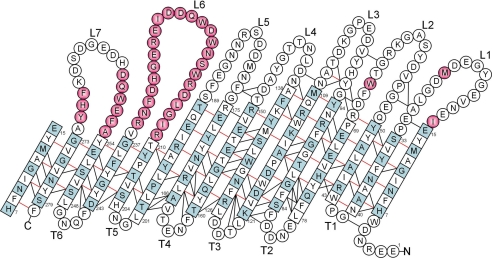
To obtain unequivocal NOE assignments, 15N-15N-1H TROSY–NOESY–TROSY and 15N-1H-1H NOESY–TROSY experiments were performed. In the TROSY–NOESY–TROSY experiment, a total of 343 backbone HN-HN NOE peaks were assigned, and 293 of them were integrated. In the NOESY–TROSY experiment, 525 backbone HN-HN NOE peaks were assigned, and 434 of them were integrated. Many strong long-range NOEs indicative of antiparallel β-sheets were present in both types of NOESY spectra. Each strong long-range HN-HN NOE was confirmed by two sets of cross-peaks, and problems of peak overlap in the integration were minimized because each NOE distance was characterized by four different NOE peaks. For weak NOEs, usually only the NOESY–TROSY peak set was identified and integrated. Altogether, we obtained a total of 316 unique backbone HN-HN distances calculated from integrated peak volumes. Among them, 137 were sequential, 46 were medium-range, and 133 were long-range NOEs (Fig. 4).
Fig. 4.
Topology map of OmpG. β-Strand residues determined by the algorithm of Kabsch/Sander (26) on the solution structures are denoted by squares, and others are denoted by circles. β-Strand residues with their side chains facing the lumen of the pore are colored in light blue. Hydrogen bond pairs observed by NOEs are indicated by red lines. Additional long-range backbone HN-HN NOEs are indicated by black lines. Unassigned residues are colored in pink. Loops (L) and turns (T) are numbered from N to C terminus.
Solution NMR Structure of OmpG in DPC Micelles.
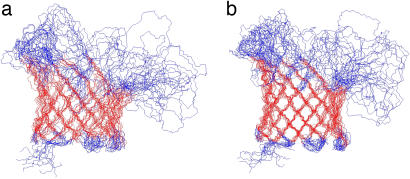
Structures calculated based on the 316 NOE and 262 dihedral angle restraints are displayed in Fig. 5a. A 14-stranded β-barrel is clearly defined by long-range NOEs observed between neighboring antiparallel β-strands. The sheer number is n + 2 = 16 (n is the number of β-strands in the barrel, and 2 is the hydrogen-bonded offset between neighboring β-strands), and the tilt angle of β-strands is 41° accordingly (24, 25). Similar to other β-barrel MPs, the side chains of the β-strands alternate in being directed toward the lipid bilayer and the lumen of the pore (Fig. 4). The only anomaly in this pattern is found at residue L78, which points toward the lipid bilayer, i.e., in the same direction as the preceding residue F77. A closer look reveals that F77 and L78 are part of a β-bulge, and in fact almost equally strong backbone HN-HN NOEs were observed for both F77-F85 and L78-F85. Overall, the β-strand residues of OmpG are well defined with a backbone root-mean-square deviation (rmsd) of 2.38 ± 0.46 Å (Table 1). The residues of the six periplasmic turns are also adequately defined, leading to a backbone rmsd of the β-strand plus turn residues of 2.54 ± 0.47 Å. However, like in other β-barrel MPs that have been studied by NMR, the extracellular loops are very flexible; the backbone rmsd is 7.37 ± 1.29 Å when the disordered loops are also included. The precision of the barrel portion of this structural ensemble is somewhat degraded by a paucity of interstrand NOEs between strands β10 and β11, and β11 and β12 (only five long-range NOEs for each). There are two proline residues in β10 and β11 strands, which disrupt the characteristic antiparallel β-strand hydrogen bonding pattern and the number of observed long-range NOEs. Therefore, strand β10 appears more flexible with different conformers adopting different degrees of bending toward the middle of the barrel. The poorer precision and relatively short length of the last few β-strands is probably also a result of almost all unassigned residues belonging to loops 6 and 7 that are connected to this region (Fig. 4). These loops probably undergo slow conformational exchange resulting in nonobservable NMR signals.
Fig. 5.
Superposition of the10 lowest-energy conformers calculated with 316 NOE distance and 262 dihedral angle restraints (a) and with the addition of 220 hydrogen bond restraints (b). Residues in red are β-sheet residues (see Table 1) that are used for superposition. All structure figures were prepared by MOLMOL (26).
Table 1.
Statistics of the OmpG solution NMR structure
| Structures calculated with NOE and dihedral angles* | Structures calculated with additional H-bond restraints† | |
|---|---|---|
| Backbone rmsd of β-sheet residues‡ | 2.38 ± 0.46 | 1.43 ± 0.30 |
| vs. PDB entry 2IWV§ | 2.44 ± 0.27 | 1.71 ± 0.25 |
| vs. PDB entry 2IWW§ | 2.46 ± 0.27 | 1.68 ± 0.22 |
| vs. PDB entry 2F1C§ | 2.46 ± 0.27 | 1.65 ± 0.22 |
| Backbone rmsd of β-sheet and turn residues¶ | 2.54 ± 0.47 | 1.67 ± 0.29 |
| Backbone rmsd of all residues | 7.37 ± 1.29 | 7.03 ± 1.25 |
| Ramachandran map analysis‖ | ||
| Most favored, % | 66.7 | 66.8 |
| Additionally allowed, % | 27.1 | 27.1 |
| Generously allowed, % | 4.2 | 4.4 |
| Disallowed, % | 1.9 | 1.6 |
*Structure calculations performed with 316 NOE upper limit distance restraints and 262 dihedral angle restraints.
†Structure calculations performed with same restraints as in * except that 220 hydrogen bond restraints were added.
‡β-sheet residues are 7–15, 33–40, 43–50, 69–78, 84–94, 109–121, 126–138, 150–160, 166–175, 190–201, 204–210, 237–243, 248–254, and 273–279.
¶β-sheet and turn residues are 7–15, 33–50, 69–94, 109–138, 150–175, 190–210, 237–254, and 273–279.
‖Analyzed by using PROCHECK-NMR (37).
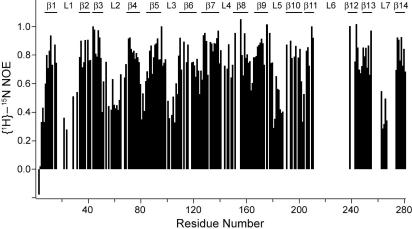
The flexibility of the loop residues was directly confirmed by {1H}-15N heteronuclear NOE measurements, which are sensitive to the mobility of individual amide N–H bond vectors (Fig. 6). High (theoretical limit 0.86 at 800 MHz) and low values indicate restricted and extended motions, respectively, on the picosecond-to-nanosecond time scale. A total of 189 residues were analyzed, and the average values were 0.83 for β-barrel, 0.63 for turn, and 0.57 for loops residues. Therefore, as observed for other β-barrel MPs (13), the barrel is quite rigid, the turns exhibit some flexibility, and the loops are more mobile.
Fig. 6.
Heteronuclear {1H}-15N NOEs of OmpG measured at 800 MHz.
Because the observed NOEs provide strong evidence for hydrogen bonds between antiparallel β-strands, hydrogen bond restraints in these positions were added in a next round of structure calculations. A total of 55 H-bond pairs were identified based on satisfying two criteria: a strong backbone HN-HN NOE was observed between each pair, and both residues of the pair exhibited clear secondary chemical shifts indicative of β-structure (Fig. 4). The inclusion of 220 additional hydrogen bond restraints in the structure calculation improved the quality of the structure (Fig. 5b), which now had a backbone rmsd of the β-barrel residues of 1.43 ± 0.30 and an rmsd of the β-barrel and turn residues of 1.67 ± 0.29 (Table 1). On average, the secondary structure determined by the Kabsch–Sander algorithm (26) yielded 131 residues participating in the β-sheet as shown in Fig. 4.
Comparison with Crystal Structures.
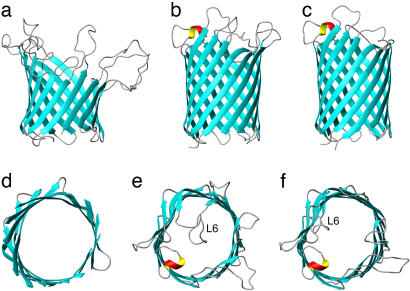
Very recently, two groups published crystal structures of OmpG. In one study, OmpG was extracted from native cell outer membranes with lauryl-N,N-dimethyldodecylamine-N-oxide (LDAO) and purified in its native form in tetraoxy-ethylene-mono-n-octylether (C8E4). Crystals were formed in C8E4 at pH 5.5 (17). This structure showed a 14-stranded β-barrel with a wide open pore with no loops folded back into the barrel lumen. Consistent with the positive secondary chemical shifts (Fig. 3), a single turn of α-helix was observed in loop 4. In the other study, OmpG was expressed without its signal sequence, extracted and purified from inclusion bodies, and subsequently refolded in detergent micelles (18). Two different crystal forms that were suitable for structure determination were obtained. At pH 7.5 in LDAO, OmpG was a 14-stranded β-barrel in an open state in which no loop residues folded back into pore lumen. However, at pH 5.6 in β-OG, OmpG was a 14-stranded β-barrel in a closed state, in which the extracellular loop 6 was folded back into the pore lumen of the barrel, thereby blocking free passage through the pore (Fig. 7). The closure of the OmpG pore apparently is caused by the rearrangement of loop 6, which is triggered by the repulsion of two protonated histidines in neighboring strands β12 and β13 at acidic pH. Surprisingly, the Subbarao and van den Berg structure resembles the open state and not the closed state of the two Kühlbrandt structures, even though the Subbarao and van den Berg crystal was formed at pH 5.5. The reasons for this discrepancy are presently unresolved. Unfortunately, most of our unassigned residues are in this interesting region, and therefore, the present NMR structure obtained at pH 6.3 does not resolve this issue. Besides the major conformational change in loop 6, loops 1, 2, and 7 are also reoriented to different degrees, whereas loops 3, 4, and 5 remain unaffected (18). Despite these differences, which may be due to pH, detergent, purification scheme, or different crystal contacts, the β-barrel regions of the three crystal structures agree very well with each other with a backbone rmsd of 0.43 ± 0.06 Å. The β-barrel backbone rmsd between the NMR solution structure and the crystal structures varies from 1.65 ± 0.22 to 1.71 ± 0.25, depending on which crystal structure is chosen (Table 1). Except for three well defined NOEs in turn 4 [and a couple of NOEs in the flexible loops and N terminus; see supporting information (SI) Data Set], all NMR distance and angle restraints were consistent with the crystal structures. One obvious difference between the barrel portions of the crystal and NMR structures is the rounder shape of the NMR structure compared with the crystal structures. At this point, it is not clear whether this difference is the result of the current somewhat limited set of experimental NMR restraints, crystal contacts in the crystal structures, different lipid environments, or a combination of these. Measurements of residual dipolar couplings should improve the precision of the NMR structure and thus resolve the question of whether these details of the crystal and solution structures are different indeed.
Fig. 7.
Comparison of solution and crystal structures of OmpG. (a and d) Lowest-energy conformer of the 10 NMR structure ensemble calculated by NOE, dihedral angle, and hydrogen bond restraints. (b and e) Crystal structure in the closed state (PDB entry 2IWW). (c and f) Crystal structure in the open state (PDB entries 2F1C and 2IWV for c and f, respectively). a–c are side views, and d–f are views of the barrel from the extracellular side. The unstructured loops and N terminus are omitted in d for a clearer view.
Although some residues in loop 4 had positive secondary chemical shift values, indicative of a short α-helix in this region, the NMR structure calculations did not pick up the formation of this α-helix. We attribute this to an insufficient number of observed NOEs in this region. Among all seven extracellular loops, L4 is the one with the largest heteronuclear NOE values, indicative of a more ordered structure (Fig. 6). The flexibility of the loop residues in OmpG leads to a shorter barrel on the extracellular side of the calculated NMR structure ensemble compared with the corresponding crystal structures. Very similar effects were observed previously when solution and crystal structures of two smaller β-barrel MPs, OmpA and PagP, were compared (3, 6). The differences may also be due to the different detergents that were used in the different studies. However, the hydrophobic lengths of DPC and LDAO are similar, but those of β-OG and C8E4 are shorter so that matching of the detergent chain length and hydrophobic barrel length does not appear to be the prime reason for the observed differences. The shorter and rounder barrel and the more disordered loops in the solution structures are the primary reasons for the larger rmsd values between the NMR and crystal structures than between the crystal structures among themselves. It is possible that these loops of OmpG (and other β-barrel MPs) are natively unfolded or interact with lipopolysaccharides in the outer membrane and that the crystal structures give an exaggerated sense of certainty about the physiological structures of these parts of the protein. It should be kept in mind that OmpG crystallized with the barrels forming long tubes or other head-to-head or head-to-tail arrangements with the loops forming many nonnative protein–protein contacts. The β-barrel of the OmpG solution structure also showed slightly higher rmsd values compared with the OmpA (and other eight-stranded β-barrel) solution structure(s), even though the average number of observed NOEs, particularly the long-range NOEs, per residue is higher in OmpG than in OmpA. The bigger size of the OmpG barrel allows for greater conformational freedom of the barrel residues without raising energy penalties.
The NMR solution structure was obtained at pH 6.3, i.e., in between the pH values at which OmpG crystallized in the open and closed forms. Conlan and Bayley (20) found that the apparent pKa for the open-to-closed transition of OmpG is ≈6.0. Therefore, we would expect that in solution, loop 6 and, to a lesser degree, loops 1, 2, and 7 are in a dynamic equilibrium between the open and closed states around pH 6. It is therefore not surprising that few NMR peaks were obtained from loop 6, which likely leads to exchange-broadened NMR signals in this region. Despite severe line-broadening, we were able to assign segments that were likely part of loops 6 and 7. However, because these assignments were only tentative, we excluded them from the present study. Future experiments under different pH and/or temperature conditions, perhaps even with some physiological lipid supplements, may shed more light on this interesting region and the possible gating mechanism of the OmpG channel.
In summary, the structure of the largest monomeric MP studied to date by solution NMR is in good agreement with crystal structures in the β-barrel region, although the barrel is a little shorter and more dynamic toward the extracellular space in the solution structure. Under the current NMR conditions, OmpG is likely present as a mixture of open and closed conformers that are in conformational exchange on the microsecond-to-millisecond time scale. Further NMR studies of OmpG at different pH values may not only elucidate the gating mechanism of this unique monomeric porin but may also help with the design of future applications of OmpG as a relatively simple engineerable conductance-based biosensor.
Materials and Methods
Expression and Purification of OmpG.
The plasmid containing the pT7-SMC vector and the sequence of mature OmpG of E. coli with a methionine replacing the signal sequence was a kind gift of Dr. H. Bayley (Oxford University, Oxford, U.K.) (16). The protein was expressed in the BL21(DE3)pLysE cells under the control of the T7 promoter. When the OD value reached 0.6, expression was induced by adding isopropyl-β-d-thiogalactoside to a final concentration of 0.5 mM. Cells were grown for another 4 h before harvesting; however, up to 7 h was allowed when cultures were grown in D2O media. For a 1-liter culture, the cell pellet was first dissolved in 10 ml of lysis buffer (200 μg/ml hen egg white lysozyme/10 mM Tris/1 mM EDTA, pH 8.0) and stirred on ice for 30 min. The lysate was French-pressed and then centrifuged at 2,600 × g for 30 min at 4°C. The OmpG-containing pellet was dissolved in 50 ml of denaturation buffer (8 M urea/10 mM Tris, pH 8.0) and passed through a sterile filter. The degassed solution was loaded onto a DEAE-Sepharose column and eluted with a 0–250 mM NaCl gradient in denaturation buffer. The purest OmpG fractions, as judged from SDS/PAGE, were concentrated to ≈15 mg/ml for the next step or storage. Final yields of pure OmpG from 1-liter culture varied from 10 to 30 mg. For uniformly 13C,15N,2H-labeled OmpG, cells were grown in D2O minimal media supplemented by 2 g/liter 13C/2H-labeled glucose, 1g/liter (15NH4)2SO4, and 10% CDN100-Bioexpress. In this case, the yield was ≈15 mg/liter.
Refolding and Sample Preparation for NMR.
Many detergents were tested under different conditions. The best refolding condition was achieved when concentrated OmpG was added dropwise to 30 ml of stirred 10 mM Tris buffer (pH 9), which contained 70 mM n-octyl-β-glucopyranoside (β-OG) (Sigma). This refolding solution was then kept at 37°C for at least 48 h. Using this protocol, we achieved a refolding efficiency of >95% as judged by SDS/PAGE without sample boiling. Samples for electrophysiological recordings in planar bilayers were taken at this stage.
Refolded OmpG in β-OG was concentrated to <1 ml by using a 30-kDa nominal-molecular-weight-limit ultrafiltration membrane (Millipore) in an Amicon concentrator. A 10-ml 15 mM DPC (Anatrace) micelle solution in 25 mM bis-Tris buffer (pH 6.3), containing 50 mM NaCl and 0.05% NaN3 was subsequently added dropwise while stirring. This solution was stirred for another 1 h at room temperature, further incubated at 4°C overnight, and concentrated by ultrafiltration. The sample was subjected to one more cycle of DPC/bis-Tris buffer exchange and concentration to give OmpG/DPC samples for triple-resonance NMR experiments with final concentrations of 1.0–1.3 mM 13C,15N,2H-labeled OmpG.
Planar Lipid Bilayer Recordings.
Planar lipid bilayers were prepared from a 15 μg/ml solution of diphytanoyl-phosphatidylcholine (Avanti Polar Lipids) in n-decane (Aldrich). The lipid solution was painted on a 500-μm-diameter hole in a Teflon partition separating two 1.5-ml compartments that were filled with 1 M KCl solutions in 10 mM Tris·Cl (pH 7.4). A chlorided silver electrode was immersed in each compartment. The front (cis) side was grounded and the rear (trans) side was connected to an Axopatch 200B amplifier (Axon Instruments). The formation of a planar lipid bilayer was tested by optical reflectance, resistance, and capacitance. One to 2 μl of 25 μM refolded OmpG in β-OG micelles was added to the front side of the preformed planar bilayer. After a few minutes of equilibration, a voltage of 40 mV was applied to the trans compartment, and the channel activity was recorded on magnetic medium.
NMR Spectroscopy.
For sample optimization, 2D-TROSY (27) spectra of 15N-labeled or 15N,2H-labeled samples were collected on a 500-MHz Varian UNITYINOVA spectrometer at different temperatures. All 3D NMR spectra of triple-labeled samples were collected at 40°C on an 800-MHz Varian INOVA spectrometer equipped with a triple-resonance cryoprobe. For sequential backbone assignments, TROSY versions of HNCA, HN(CO)CA, HN(CA)CB, HN(COCA)CB, HNCO, and HN(CA)CO experiments were collected (28). A new, just-in-time (JIT) version of HN(CA)CO (29) was critical to obtaining the backbone CO assignments. 15N-15N-1H TROSY–NOESY–TROSY (P. Zhou, personal communication), and 15N-1H-1H NOESY–TROSY (30) experiments were performed with mixing times of 200 ms. TROSY-based 2D 1H-15N heteronuclear NOE experiment was also performed at 800 MHz with a saturation delay of 5 s (31). Spectra were processed and analyzed with NMRPipe (32) and Sparky (33), and all indirect dimensions in the 3D spectra were processed with forward–backward linear predictions.
Structure Calculations.
Backbone dihedral angle restraints were obtained by using the TALOS program (34) with backbone chemical shifts that were calibrated for isotope and TROSY effects (35). Predictions of φ and ψ dihedral angles from the program were used in the structure calculations as input restraints with ±30° error tolerances. Assigned NOE peaks were individually integrated with Gaussian lineshapes and converted to distances that were calibrated based on the fact that the average HN-HN distance between antiparallel β-strands is 3.3 Å. For structure calculations, the distances were categorized as having strong, medium, and weak NOEs with upper limits of 3.5, 5.0, and 6.0 Å, respectively. When used, hydrogen bonds were each restrained with two upper limits of 2.5 and 3.5 Å for HN···O and N···O, respectively. All structure calculations were performed by using the CNS v.1.1 on a Linux cluster (36). Starting from an extended structure, 4,000 steps of high-temperature annealing, 8,000 steps of torsion-angle slow-cooling annealing, and 8,000 steps of Cartesian slow-cooling annealing were used to fold the protein. Ten structures with the lowest overall violation energies were selected from 100 calculated conformers for the final representation of each calculation.
Supplementary Material
Acknowledgments
We thank Dr. H. Bayley for the OmpG plasmid; Dr. P. Zhou (Duke University, Durham, NC), Dr. R. A. Venters (Duke University), Mr. J. W. Werner-Allen (Duke University), and Dr. J. H. Bushweller (University of Virginia) for NMR pulse sequences and helpful comments; and Dr. J. Schmittschmitt and Mr. W. Miller for preliminary studies on OmpG. This work was supported by National Institutes of Health Grant GM051329.
Abbreviations
- DPC
dodecylphosphocholine
- MP
membrane protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates of the OmpG structure have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2JQY).
This article contains supporting information online at www.pnas.org/cgi/content/full/0705466104/DC1.
References
- 1.White SH. 2007 http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html.
- 2.Sprangers R, Kay LE. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 3.Arora A, Abildgaard F, Bushweller JH, Tamm LK. Nat Struct Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 4.Fernández C, Adeishvili K, Wüthrich K. Proc Natl Acad Sci USA. 2001;98:2358–2363. doi: 10.1073/pnas.051629298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández C, Hilty C, Wider G, Güntert P, Wüthrich K. J Mol Biol. 2004;336:1211–1221. doi: 10.1016/j.jmb.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CR, Prive GG, Bishop RE, Kay LE. Proc Natl Acad Sci USA. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang PM, Bishop RE, Kay LE. Proc Natl Acad Sci USA. 2004;101:9618–9623. doi: 10.1073/pnas.0402324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oxenoid K, Chou JJ. Proc Natl Acad Sci USA. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chill JH, Louis JM, Miller C, Bax A. Protein Sci. 2006;15:684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arora A, Tamm LK. Curr Opin Struct Biol. 2001;11:540–547. doi: 10.1016/s0959-440x(00)00246-3. [DOI] [PubMed] [Google Scholar]
- 11.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. J Biomol NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 12.Sanders CR, Sönnichsen F. Magn Reson Chem. 2006;44:S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 13.Tamm LK, Liang B. Prog NMR Spectrosc. 2006;48:201–210. [Google Scholar]
- 14.Fajardo DA, Cheung J, Ito C, Sugawara E, Nikaido H, Misra R. J Bacteriol. 1998;180:4452–4459. doi: 10.1128/jb.180.17.4452-4459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misra R, Benson SA. J Bacteriol. 1989;171:4105–4111. doi: 10.1128/jb.171.8.4105-4111.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conlan S, Zhang Y, Cheley S, Bayley H. Biochemistry. 2000;39:11845–11854. doi: 10.1021/bi001065h. [DOI] [PubMed] [Google Scholar]
- 17.Subbarao GV, van den Berg B. J Mol Biol. 2006;360:750–759. doi: 10.1016/j.jmb.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 18.Yildiz O, Vinothkumar KR, Goswami P, Kühlbrandt W. EMBO J. 2006;25:3702–3713. doi: 10.1038/sj.emboj.7601237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamm LK, Arora A, Kleinschmidt JH. J Biol Chem. 2001;276:32399–32402. doi: 10.1074/jbc.R100021200. [DOI] [PubMed] [Google Scholar]
- 20.Conlan S, Bayley H. Biochemistry. 2003;42:9453–9465. doi: 10.1021/bi0344228. [DOI] [PubMed] [Google Scholar]
- 21.Kleinschmidt JH, Wiener MC, Tamm LK. Protein Sci. 1999;8:2065–2071. doi: 10.1110/ps.8.10.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong H, Szabo G, Tamm LK. Nat Chem Biol. 2006;2:627–635. doi: 10.1038/nchembio827. [DOI] [PubMed] [Google Scholar]
- 23.Metzler WJ, Constantine KL, Friedrichs MS, Bell AJ, Ernst EG, Lavoie TB, Mueller L. Biochemistry. 1993;32:13818–13829. doi: 10.1021/bi00213a010. [DOI] [PubMed] [Google Scholar]
- 24.Liu WM. J Mol Biol. 1998;275:541–545. doi: 10.1006/jmbi.1997.1501. [DOI] [PubMed] [Google Scholar]
- 25.Schulz GE. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 26.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 27.Pervushin K, Riek R, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang D, Kay LE. J Am Chem Soc. 1999;121:2571–2575. [Google Scholar]
- 29.Werner-Allen JW, Jiang L, Zhou P. J Magn Reson. 2006;181:177–180. doi: 10.1016/j.jmr.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Zhu G, Kong XM, Sze KH. J Biomol NMR. 1999;13:77–81. doi: 10.1023/A:1008398227519. [DOI] [PubMed] [Google Scholar]
- 31.Zhu G, Xia Y, Nicholson LK, Sze KH. J Magn Reson. 2000;143:423–426. doi: 10.1006/jmre.2000.2022. [DOI] [PubMed] [Google Scholar]
- 32.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Goddard TD, Kneller DG. SPARKY 3. San Francisco: Univ of California; [Google Scholar]
- 34.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 35.Moseley HN, Sahota G, Montelione GT. J Biomol NMR. 2004;28:341–355. doi: 10.1023/B:JNMR.0000015420.44364.06. [DOI] [PubMed] [Google Scholar]
- 36.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.