Abstract
Side-population (SP) cells within cancers and cell lines are rare cell populations known to enrich cancer stem-like cells. In this study, we characterized SP cells from the human breast cancer cell line MCF7 as a model for cancer stem-like cells. Compared with non-SP cells, MCF7 SP cells had higher colony-formation ability in vitro and greater tumorigenicity in vivo, suggesting that MCF7 SP cells enrich cancer stem-like cells. cDNA microarray analysis of the SP cells indicated higher expression of ATP-binding cassette transporters and genes involved in quiescence, which were confirmed by quantitative RT-PCR and flow cytometry cell cycle analysis. To identify signal pathways important for cancer stem-like cells, we analyzed cDNA microarray data and identified nine pathways that were altered in the SP cells. To analyze the protein signaling networks, we used reverse-phase signaling pathway protein microarray technology and identified three signaling proteins that are significantly different between MCF7 SP and non-SP cells. Notably, signaling of phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR), signal transduction and activator of transcription (STAT3), and phosphatase and tensin homolog (PTEN) was confirmed to be critical for MCF7 SP cell survival and proliferation by pathway specific inhibitors, selected gene knockdown, and in vivo tumorigenicity assay. The STAT3 pathway was found to be positively regulated by mTOR signaling, whereas PTEN served as a negative regulator of both STAT3 and mTOR signaling. This study suggests the existence of prosurvival signaling pathways critical for cancer stem-like cell maintenance, which could be selectively targeted for inhibiting cancer stem-like cells for improved treatment.
Keywords: cDNA microarray, reverse-phase protein array, side population cells, signaling pathway
Cancer stem cells, which are defined as cells capable of giving rise to a new tumor, are thought to be the root cause of cancer. Cancer stem cells were first identified in acute myelogenous leukemia (1). Almost 10 years later, the first solid tumor stem cells were identified in breast cancer when it was demonstrated that a subpopulation of cells expressing the CD44+CD24− phenotype could form tumors with as few as 100 cells, whereas tens of thousands of the bulk tumor cells did not do so (2). Since then, cancer stem cells have been characterized in human brain tumors (3) and colon (4, 5), head and neck (6), and pancreatic cancers (7).
Although most cancer stem cells were initially identified in primary patient samples, some cancer cell lines were also shown to harbor cancer stem-like cells (8–10). Cancer stem-like cells from cell lines could be a promising model for cancer stem cell research because of their unlimited supply and ease of handling. Accumulating evidence suggests that side-population (SP) cells, a small population of cells from cancer cell lines, are enriched in a subset of cancer stem-like cells (8–10). SP cells were first defined by Goodell et al. (11) in the hematopoietic system. Although the mechanism for producing the SP phenotype is unclear, it is believed that certain ATP-binding cassette (ABC) transporters, including ABCG2/BCRP, ABCB1/MDR1, and ABCA3, which can pump out the fluorescent dye Hoechst 33342, or the relative quiescence of the cancer stem cells that limit the intake of the dye, may cause the SP phenotype (10, 12). Based on the lack of accumulation of the Hoechst dye, SP cells can be isolated by flow cytometry and have proved to enrich cancer stem-like cells in various tumors or tumor cell lines (8–10).
Breast cancer is the most frequent malignancy among women, with an incidence rate in the United States of 111 cases per 100,000 woman-years (wy) and a mortality rate of 24 deaths per 100,000 wy (13). Most recent studies indicate that breast cancer is caused by cancer stem cells, and the cure of breast cancer requires eradication of breast cancer stem cells (2, 14–16). However, the cell signaling pathways that are unique for breast cancer stem cells are not well defined. Here, using SP cells from the human breast cancer cell line MCF7 as a model of breast cancer stem-like cells, we isolated and characterized the SP cells and determined the gene expression profile of the SP cells by cDNA microarray and reverse-phase protein array (RPA) and identified signaling pathways that are preferentially activated in SP over non-SP cells. The phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR), signal transduction and activator of transcription (STAT3), and phosphatase and tensin homolog (PTEN) signaling, which form a complex signaling network, were found to be important for SP cell survival and proliferation. This study may have implications for the development of agents that target cancer stem-like cell populations for improved treatment of cancer.
Results
Existence of SP Cells in the MCF7 Cell Line.
Consistent with the findings of other groups (8, 9), the MCF7 cell line was found to contain SP cells, which presented as a distinct “tail” in the flow cytometry histogram [supporting information (SI) Fig. 3A]. The SP population could be blocked by known ABC transporter inhibitors, including verapamil, reserpine, and fumitremorgin C (FTC). As shown in SI Fig. 3A, 50 μM verapamil, 20 μM reserpine, and 10 μM FTC significantly decreased the SP population from 1.21 ± 0.38% in the control to 0.10 ± 0.03% (P = 0.06), 0.02 ± 0.01% (P = 0.05), and 0.04 ± 0.02% (P = 0.05), respectively.
SP Cells Have Higher Colony-Formation Ability in Vitro and Tumorigenicity in Vivo.
To provide evidence that the SP cells in MCF7 cells were enriched in cancer stem-like cells, we determined the tumorigenic potential of the SP and non-SP cells in vitro and in vivo. As shown in SI Fig. 3B, the SP cells had 13-fold higher colony-formation ability than the non-SP cells. With the inoculation of 5,000 cells, the SP cells formed 298 colonies in soft agar (colony-formation efficiency was 6.0%), compared with 23 for the non-SP cells (colony-formation efficiency was 0.5%) (SI Fig. 3B). The higher colony-formation ability of the SP cells suggests that SP cells may have higher tumorigenicity in vivo. To prove this, tumorigenicity assay was performed by injecting both the SP and non-SP cells into NOD/SCID mice with estrogen supplement. The SP cells showed significantly higher tumorigenicity than the non-SP cells (Table 1). Seven of 10, 11 of 12, and 7 of 8 mice inoculated with 2,000, 20,000, and 200,000 SP cells gave rise to tumors, whereas only 2 of 10, 3 of 12, and 3 of 8 inoculations of the same number of the non-SP cells grew tumors smaller than those from the SP cells (Table 1). Histological analysis indicates that the SP cell and the bulk MCF7 cell-derived tumors showed typical pathological features of cancer with many poorly differentiated round cells (SI Fig. 3C), whereas the majority of non-SP cell-derived tumors had a different pathology profile with more differentiated cells and fewer poorly differentiated cells (SI Fig. 3C). The higher colony-formation ability in vitro and higher tumorigenicity in vivo of SP cells suggest that MCF7 SP cells were enriched in cancer stem-like cells.
Table 1.
Tumorigenicity assay of MCF7 SP cells in NOD/SCID mice
| Cell number for injection |
|||
|---|---|---|---|
| 2 × 105 | 2 × 104 | 2 × 103 | |
| SP | 7/8 | 11/12 | 7/10 |
| Non-SP | 3/8 | 3/12 | 2/10 |
| P value | 0.01 | 0.06 | 0.02 |
To determine the stability of the SP cells, we sorted out the SP cells and cultured them in vitro. The cultured SP cells were restained with Hoechst dye at 4 and 18 days after culture and were reanalyzed by flow cytometry. Our data indicated that the SP fraction rapidly underwent a symmetric division and generated both SP and non-SP cells. The SP fraction in 4- and 18-day cultures was 22.3% and 5.83%, respectively. In contrast, the sorted non-SP cells were not able to generate SP cells (<0.05%) in this period of culture.
Molecular Characterization of the SP Cells.
To identify the genes that are preferentially expressed in the SP compared with the non-SP cells, microarray analysis of the gene expression profile of the two cell types was performed and compared. The flow cytometric gating used to sort SP and non-SP cells was indicated by trapezoids in SI Fig. 4. In total, 179 genes were identified, where 174 were up-regulated by >1.5-fold, and five were down-regulated by >2-fold. Thirteen genes were picked (SI Table 4) for quantitative RT-PCR confirmation, and all had similar gene expression patterns as the microarray. All 179 genes with annotations are presented in SI Table 5. Among the most significant changes are genes involved in the multidrug-resistant (MDR) drug transporters, cell cycle regulation, and signaling pathways, which will be discussed separately below.
MDR Drug Transporters.
We found higher expression of the BCRP/ABCG2 gene by both microarray (6.71-fold) and quantitative RT-PCR (7.2- ± 1.3-fold). Similarly, expression of MDR1/ABCB1 was also increased in the SP cells (11.1- ± 2.5-fold). The higher expression of both BCRP/ABCG2 and MDR1/ABCB1 suggested these transporters play important roles for the SP phenotype. It was also known that overexpression of either BCRP/ABCG2 and MDR1/ABCB1 was one of the common mechanisms of drug resistance (17). To test the drug sensitivity of the SP cells, both SP and non-SP cells were sorted into 96-well plates and treated with the cancer drugs mitoxantrone and carboplatin. As shown in SI Fig. 5 A and B, the SP cells were more resistant than the non-SP cells to mitoxantrone and carboplatin. Reserpine, which blocks the SP phenotype as shown in SI Fig. 3A, partially reversed the resistance of the SP cells to both mitoxantrone and carboplatin (SI Fig. 5 A and B). Mitoxantrone at the concentration of 50 nM inhibited the non-SP cells by 41.1 ± 0.6% but inhibited the SP cells by 22.2 ± 2.9%. When combined with repserpine (5 μM), 50 nM mitoxantrone could inhibit SP cell growth by 31.5 ± 2.2%.
Cell Cycle Regulation.
Three genes, EXT1, INHBA, and CCNT2, were identified by the microarray to be related to cell cycle regulation. In particular, both EXT1 and INHBA, which are negative cell cycle regulators, were up-regulated by 4.1- and 2.1-fold, respectively. This suggested that the SP cells contain a more quiescent population than the non-SP cells. This finding was confirmed by flow cytometry cell cycle analysis (SI Fig. 6). Compared with the non-SP cells, which contained 59% G1/G0 phase cells, the SP cells contained a higher percentage of G1/G0 phase cells (78%). Approximately 22% and 8% of the SP cells were in S and G2 phase, respectively, compared with 31% and 13% for the non-SP cells. This finding suggests that the SP cells contain larger percentage of quiescent population than non-SP cells, which may be one of the reasons for the drug resistance of the SP cells in addition to the overexpression of the ABC transporters, as shown above.
Signaling Pathways of the SP Cells.
To shed light on the SP-specific signaling pathways, all 179 genes identified in the microarray were subjected to pathway analysis using Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA). Sixty-nine genes [68 were up-regulated and one (cytochrome c) down-regulated] were mapped in the Ingenuity database and assigned to genetic networks. Genes belonging to nine signaling pathways, including the PI3K/AKT, JAK/STAT, MAPK/ERK, TGF-β, estrogen receptor, VEGF, ErbB/EGF, NF-κB, and Wnt/b-catenin pathways, were found to preferentially expressed in SP compared with non-SP cells (SI Table 6).
Identification of Signaling Pathway Proteins by Reverse-Phase Protein Array.
Signaling pathway information identified by the microarray has certain limitations, because it detects changes only in mRNA levels but cannot predict the phosphorylation status of the signaling proteins. To overcome this potential drawback, we further explored the differentially activated signaling pathway proteins of the SP cells by reverse-phase signaling pathway protein array (RPA) using a suite of phosphor-specific antibodies. Thirty-four extensively validated antibodies (by Western blot and peptide competition), which cover the signal pathways identified in microarray, were selected (SI Table 7). Three phosphorylated proteins were found to be significantly different between the SP and non-SP cells (SI Fig. 5C). The STAT protein pSTAT3 727 was significantly up-regulated in the SP cells by 1.8-fold (P < 0.0625). Differential activation/phosphorylation of p90RSK S380 and pMEK1/2 S217/221 between the SP and non-SP cells was also identified (SI Fig. 5C). p90RSK was found to be down-regulated by 2.1-fold and pMEK1/2 S217/221 increased by 1.7-fold (P < 0.0625) (SI Fig. 5C). The differential phosphorylation levels suggest that the related signaling pathways may be important for the SP cells.
Role of mTOR Signaling in SP Cell Proliferation and Survival.
As shown in SI Table 6, five genes, including PIK3C2A, PIK3R1, SOS1, SOS2, and FKHR, were mapped to the PI3K pathway. PIK3C2A, PIK3R1, SOS1, and SOS2 were chosen for further confirmation by RT-PCR. Consistent with our microarray data, all four genes, including PIK3CA2, PI3R1, SOS1, and SOS2, were expressed in the SP cells at higher levels than the non-SP cells by 1.6 ± 0.2-, 3.4 ± 0.2-, 1.9 ± 0.4-, and 7.3 ± 1.0-fold, respectively (SI Table 4). These data suggest that the PI3K/mTOR pathway may be important for SP cell survival. To confirm this, LY294002, a PI3K-specific inhibitor, and rapamycin, a specific inhibitor of mTOR, were tested for their ability to decrease the SP fraction in MCF7 cells. The MCF7 cells were treated with LY294002 and rapamycin at 2.5 and 5 μM, respectively, for 3 days and subjected to SP analysis. Both inhibitors decreased the SP fraction within the MCF7 cells. As shown in Fig. 1A, compared with control MCF7 cells, which contained 1.21 ± 0.38% SP cells, MCF7 cells treated with LY294002 and rapamycin had only 0.10 ± 0.05% (P = 0.05) and 0.30 ± 0.11% (P = 0.01) SP cells, respectively. The SP fraction in inhibitor-treated cells did not recover when the cells were further cultured in fresh medium without inhibitors for at least 24 h (SI Fig. 7). The ability of the two inhibitors to decrease the SP fraction was at least partially because of their ability to preferentially inhibit the SP cells. As shown in Fig. 1B, both LY294002 and rapamycin preferentially inhibited the SP rather than non-SP cells. LY294002 (2.5 μM) and rapamycin (5 μM) inhibited MCF7 SP cell proliferation by 50.6% and 45.78%, respectively (Fig. 1B), but inhibited the non-SP cell proliferation by 22.2% and 18.3%, respectively. In addition to proliferation, LY294002 and rapamycin also preferentially inhibited colony-formation ability of the SP cells by 44.2% and 58.2% and that of the non-SP cells by 17.6% and 8.8%, respectively (Fig. 1C). However, LY294002 and rapamycin did not cause significant apoptosis on the SP cells at tested doses (SI Fig. 8), indicating that their action on the SP cells is not through apoptosis.
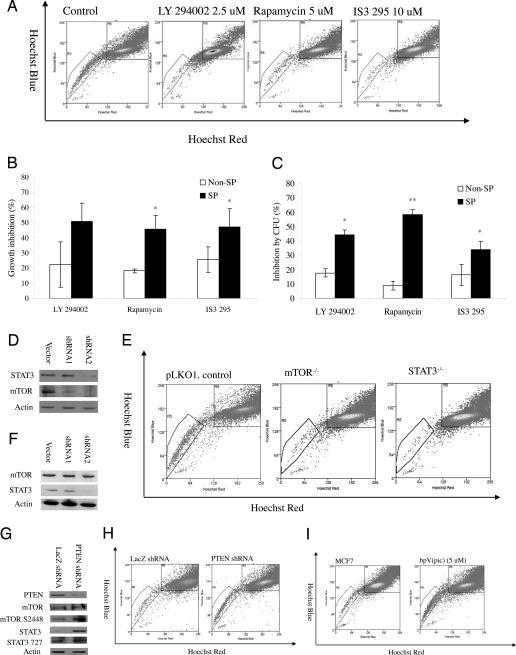
Fig. 1.
Importance of PI3K/mTOR signaling and STAT3 and PTEN signaling for SP cells. (A) Decrease of the SP fraction within MCF7 cells by pathway-specific inhibitors. (B) Proliferation inhibition effects of LY294002 (2.5 μM), rapamycin (5 μM), and IS3 295 (50 μM) on MCF7 SP and non-SP cells. (C) Colony-formation inhibition effects of LY294002 (2.5 μM), rapamycin (5 μM), and IS3 295 (25 μM) on MCF7 SP and non-SP cells. (D) Western blot analysis of expression of STAT3 and mTOR in mTOR knockdown cells. (E) Decrease of SP fraction within stable mTOR and STAT3 knockdown MCF7 cells. (F) Western blot analysis of expression of STAT3 and mTOR in STAT3 knockdown cells. (G) Decrease of the SP fraction within MCF7 cells by pathway specific inhibitors bpV(pic). (H) Decrease of SP fraction within PTEN knockdown MCF7 cells. (I) Decrease of the SP fraction within MCF7 cells by PTEN specific inhibitors bpV(pic).
The importance of the PI3K pathway for the SP cells was further confirmed by gene knockdown experiment. We chose to silence mTOR by a lentivirus-based short-hairpin RNA (shRNA), because the inhibitor of mTOR, rapamycin, decreased SP cells in MCF7 cells (Fig. 1A). Two constructs, which are designed to silence two different targeting sequences of mTOR, were selected. Fig. 1D showed that both constructs, especially, the second construct, produced greater silencing effects. Stable cells were selected and subjected to SP analysis. As expected, there was a loss of SP fraction within the stably selected mTOR knockdown cells MCF7 mTOR−/− (mTOR-shRNA-2). MCF7 mTOR−/− cells contained only 0.27 ± 0.09% SP cells, in contrast to 1.68 ± 0.42% SP cells in the vector control cells (Fig. 1E).
Role of STAT3 Signaling in SP Cell Proliferation and Survival.
As shown in SI Table 6, cDNA microarray revealed that six genes within the JAK/STAT pathway were up-regulated in the SP cells. In addition, the expression of pSTAT3 S727 was found to significant increase by 1.8-fold by RPA (SI Fig. 5C). These data suggest the STAT3 signaling pathway is important for SP cells. To confirm the functional significance of this observation, MCF7 cells were exposed to IS3 295, a STAT3-specific inhibitor (18). Exposure of the cells to this inhibitor significantly decreased the SP fraction within MCF7 cells. Compared with parental MCF7 cells, which contained 1.21 ± 0.38% SP cells, MCF7 treated with IS3 295 (10 μM) for 3 days contained only 0.29 ± 0.2% (P = 0.06) SP cells (Fig. 1A).
The SP fraction in inhibitor-treated cells did not recover when the cells were further cultured in fresh medium without IS3 295 for at least 24 h (SI Fig. 7).
Similar to LY294002 and rapamycin, IS3 295 preferentially inhibited SP proliferation and colony formation (Fig. 1 B and C) but slightly induced apoptosis in the SP cells at tested concentrations (SI Fig. 8). IS3 295 inhibited SP cell proliferation by 47.1% and colony formation by 33.8% compared with 25.5% and 16.2% for non-SP cells (Fig. 1 B and C). Similar to the effect of mTOR gene knockdown, STAT3 knockdown cells also caused loss of the SP fraction. As shown in Fig. 1F, MCF7 cells infected by lentivirus STAT3 knockdown construct 2 gave better silencing effect than the vector control. The stable cell line MCF7 STAT3−/− (MCF7 STAT3-shRNA-2) generated from this construct contained only 0.14 ± 0.09% SP fraction, compared with 1.68 ± 0.42% for the vector control cells (P = 0.01) (Fig. 1E).
mTOR/STAT3 Signaling Pathways Are Important for Tumorigenicity.
Because high tumorigenicity in vivo is one of the hallmarks of cancer stem cells, it was necessary to determine whether the mTOR and STAT3 pathways play a role in MCF7 tumorigenicity in the mouse model. To test this, the serial number of MCF7 vector control, MCF7 mTOR−/−, and STAT3−/− knockdown cells was injected into nude mice with estrogen supplement. As expected, both mTOR−/− and STAT3−/− knockdown cells had significantly reduced ability to generate tumors in mice (Table 2). In both 1.5 × 105 and 1.5 × 104 inoculation groups, three of five mice inoculated with MCF7 vector control cells generated new tumors, whereas none of five mice in both MCF7 mTOR−/− and STAT3−/− cells knockdown groups gave rise to new tumors. Similar results were obtained for the 1.5 × 106 inoculation groups, where four of five MCF7 vector control mice had new tumors generated, but only two of five and one of five mice inoculated with MCF7 mTOR−/− and STAT3−/− had tumors 6 weeks after inoculation.
Table 2.
Tumorigenicity assay of stable mTOR and STAT3 knockdown cells in nude mice
| Cell number for injection |
|||
|---|---|---|---|
| 1.5 × 106 | 1.5 × 105 | 1.5 × 104 | |
| MCF7 vector | 4/5 | 3/5 | 3/5 |
| STAT3−/− | 1/5 | 0/5 | 0/5 |
| mTOR−/− | 2/5 | 0/5 | 0/5 |
We also determined whether the pathway-specific inhibitors could reduce tumorigenicity of MCF7 cells in mice. Indeed, when treated with all three pathway inhibitors, including mTOR inhibitor rapamycin (2.5 μM), PI3K inhibitor LY294002 (5 μM), and STAT3 inhibitor IS3 295 (10 μM) for 3 days, MCF7 cells showed decreased tumorigenicity. Five of five and four of five nude mice grew new tumors 6 weeks after inoculation of 1.5 × 106 and 1.5 × 105 MCF7 cells, whereas only two of five and one of five for mice that received LY294002 treated cells, two of five and two of five for mice that received rapamycin-treated cells, and two of five and two of five for mice that received IS3 295-treated cells generated tumors (Table 3).
Table 3.
Tumorigenicity assay of MCF7 cells treated with inhibitors in nude mice
| Cell number for injection |
||
|---|---|---|
| 1.5 × 106 | 1.5 × 105 | |
| Control cells | 5/5 | 4/5 |
| Rapamycin-treated cells | 2/5 | 2/5 |
| LY 294002-treated cells | 2/5 | 1/5 |
| IS3 295-treated cells | 2/5 | 2/5 |
MCF7 cells were treated with LY294002 (2.5 μM), rapamycin (5 μM), and IS3 295 (10 μM) for 3 days.
Regulation of STAT3 by mTOR.
Because both PI3K/mTOR and STAT3 signaling was found to be important for MCF7 SP cells (Fig. 1 A–E; Tables 2 and 3), we next explored whether the two pathways were linked (19). Western blot experiment was used to detect the change of mTOR expression in MCF7 STAT3−/− cells and to analyze the change of STAT3 expression in MCF7 mTOR−/− cells. The expression of mTOR was not altered in MCF7 cells transfected with lentivirus-based shRNAs targeting the STAT3 gene (Fig. 1F). However, the expression of STAT3 was found to decrease in MCF7 cells transfected with lentivirus-based shRNAs targeting the mTOR gene (Fig. 1D). These experiments suggest that STAT3 may be positively regulated by mTOR.
PTEN, a Negative Regulator of mTOR/STAT3 Pathway, Plays an Important Role in SP Cell Maintenance.
PTEN, a tumor suppressor, usually functions as a negative regulator of the PI3K/mTOR pathway (20, 21), which positively regulates STAT3 signaling (19). Recent studies indicate that PTEN plays an important role not only in self-renewal and activation of hematopoietic stem cells but also in the prevention of leukemogenesis (20, 21). It is quite likely that PTEN also plays an important role in breast cancer stem cells by negatively regulating PI3K/mTOR/STAT3 signaling. When PTEN was knocked down by a validated shRNA (22), expression of mTOR, pmTOR S2448, STAT3, and pSTAT3 S727 was increased (Fig. 1G). To determine whether PTEN also plays a role in SP phenotype regulation, we analyzed the SP fraction within PTEN knockdown MCF7 cells. As shown in Fig. 1H, the SP fraction increased when PTEN was knocked down. The PTEN knockdown cells contained 1.76 ± 0.25% SP cells, compared with 0.85 ± 0.10% in MCF7 control cells. The negative regulation of the SP phenotype by PTEN was further confirmed by inhibitor study. When the MCF7 cells were treated with PTEN-specific inhibitor bpV(pic) (5 μM) for 3 days, the SP fraction within the MCF7 cells was increased to 7.60 ± 0.54% compared with 1.21 ± 0.38% in the control cells (P = 0.03) (Fig. 1I). To determine whether the negative regulation of PTEN on the SP phenotype reflects the cancer stem-like cell properties, PTEN overexpressing MCF7 cells, in which PTEN was delivered by using adenovirus as reported (23), were inoculated into NOD/SCID mice, and tumor growth was monitored. Indeed, PTEN overexpression caused decreased tumorigenicity. Five of five and four of five mice grew tumors 6 weeks after inoculation of 150,000 and 15,000 vector control cells, whereas only two of five and two of five mice that received PTEN-overexpressing MCF7 cells grew tumors. These data suggest that PTEN functions as a negative regulator of mTOR and STAT3 signaling and plays an important role in SP cell maintenance within the entire population.
Discussion
Establishing an appropriate in vitro cancer stem cell model is critical for the study of cancer stem cell biology, because of limited supply of cancer stem cells from patient samples. In this study, a small fraction of SP cells isolated by flow cytometry from the human breast cancer cell line MCF7 were found to have significantly higher colony-formation ability in vitro and also higher in vivo tumorigenicity in NOD/SCID mice than the non-SP cells. These findings suggest that the SP cells are enriched in cancer stem-like cells, which is consistent with other studies on SP cells (8–10).
One potential concern for claiming the cancer stem cell-like property of the SP cells is that Hoechst 33342 dye may create bias by selectively injuring non-SP cells. To provide evidence that Hoechst 33342 dye used in the staining procedure does not directly cause the difference in “stemness” between SP and non-SP cells, we prepared a bulk control group, in which MCF7 cells were not stained with the Hoechst 33342 dye but directly sorted in comparison with non-SP cells isolated from cells stained with Hoechst 33342 dye for in vitro colony-formation assay and tumorigenicity assay. We did not observe a significant difference between the non-SP cell group and bulk cell group in both colony-formation and mouse tumorigenicity assays. With the inoculation of 5,000 cells, the non-SP cells formed 23 ± 1.5 and the unstained bulk control cells 26 ± 1.5 colonies in the colony-formation assay. In the tumorigenicity experiment, we observed that three of eight and two of eight NOD/SCID mice inoculated with non-SP cells at numbers of 200,000 and 20,000 generated tumor 6 weeks after inoculation, whereas four of eight and two of eight NOD/SCID mice inoculated with bulk unstained cells grew tumors.
cDNA microarray analysis was performed to shed light on the gene expression profile specific to SP cells. Microarray analysis identified increased expression of ABC transporter genes ABCG2/BCRP, responsible for SP phenotype in different cell types (10, 12), and the negative cell cycle regulator genes, including EXT1 and INHBA. These findings suggest that the SP phenotype could be due to either higher expression of ABC transporters or relative quiescence of the cells because of the expression of the negative cell cycle regulators, or both. Thus the SP cells may contain a larger population of quiescent than non-SP cells, which was confirmed by flow cytometry analysis, as well as cells that overexpress ABC transporters. It is quite likely that both high expression of ABC transporters and quiescent state contribute to the resistance of SP cells to clinical drugs, as shown in SI Fig. 5 A and B. We noted that the drug-resistance phenotype of the SP cells might be underestimated in this study, because SP cells could potentially “differentiate” into non-SP cells, which are more sensitive to drugs. The drug-sensitivity experiment comparing SP and non-SP cells used in this study lasted 4 days. We found that, indeed, SP cells cultured in this time period could “differentiate” into SP and non-SP cells, with only 22.3% SP cells remaining in the 4-day-old SP cell culture.
Microarray analysis of the signaling pathway genes identified nine pathways, including the PI3K/AKT, JAK/STAT, MAPK/ERK, TGF-β, estrogen receptor, VEGF, ErbB/EGF, NF-κB, and Wnt/b-Catenin pathways, that are altered in the SP compared with non-SP cells (SI Table 6). It is interesting to note that the Wnt and STAT pathways have been identified in the hepatocellular carcinoma SP cells (24) and NF-κB and MAPK pathways in tumorigenicity breast cancer cells (25). Because the cDNA array detects only mRNA changes, whereas cell signaling is mediated by posttranslational modification of proteins such as phosphorylation, we used the RPA technology (26). This technology, using extensively validated phosphor-specific antibodies, can measure and quantify dozens to hundreds of specific signaling events from only a few thousand cells, which makes this technology a powerful tool for signaling pathway analysis (26, 27). The RPA technology, which has extreme sensitivity and high-throughput capability, is ideally suited for cancer stem cell study, because cancer stem cells are a rare population within tumors, which limit the samples for routine protein detection by techniques such as Western blot (26, 27). To reduce possible false-positive results, stringent criteria were applied for RPA experimental design and data analysis. Both SP and non-SP cells were interdependently prepared four separate times. Using this stringent criterion, we identified three phosphorylated isoforms of the following proteins: pSTAT3 S727, p90RSK S380, and pMEK1/2 S217/221, which were found to be significantly different between MCF7 SP and non-SP cells. These active forms of proteins fall into two major signal pathways, JAK/STAT and ERK/MAPK, suggesting these pathways may be important for MCF7 SP cells.
To confirm the importance of the signal pathways identified in this study for the SP cell phenotype and “stemness,” we focused on the PI3K/AKT and STAT3 pathways. Inhibition of the PI3K/mTOR pathway by the specific pathway inhibitors LY294002 and rapamycin and knockdown of mTOR gene by shRNA significantly reduced not only the SP fraction within MCF7 cells but also tumorigenicity (Fig. 1 A–F and Tables 2 and 3). These findings suggest the PI3K/mTOR pathway is important for SP cell survival, and that the reduced tumorigenicity is most likely due to the decrease of cancer stem-like cells in the SP fraction. These findings are consistent with the previous observation that the PI3K/AKT pathway is important for acute myeloid leukemia stem cells (28). Similarly, treatment with the STAT3 inhibitor IS3 295 or knockdown of the STAT3 gene by shRNA caused loss not only of SP fraction but also of tumorigenicity. The decrease of tumorigenicity was consistent with reports by other groups in different tumor cells by knockdown STAT3 expression using RNAi (29) or by abrogating STAT3 (30). Interestingly, we have found that both mTOR and STAT3 are also important for SP phenotype and tumorigenicity in human lung cancer cell line A549 cells (data not shown), suggesting the importance of mTOR and STAT3 signaling is not restricted to breast cancer stem-like cells.
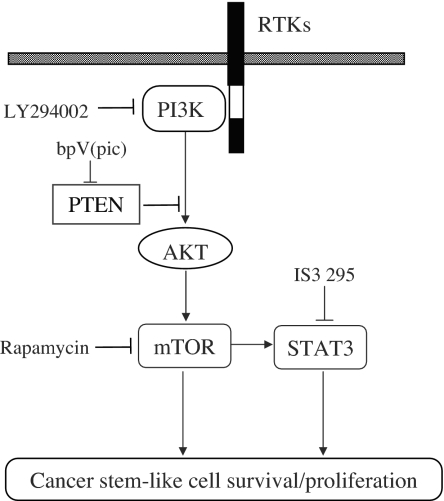
STAT3 signaling is positively regulated by PI3K/mTOR signaling (19), which is negatively regulated by PTEN (20, 21). However, the role of PTEN in the SP cell phenotype and “stemness” is unclear. Therefore, we evaluated the role of PTEN in the regulation of cancer stem-like cells. Indeed, when PTEN was inhibited by shRNA or specific inhibitor bpV(pic), the SP fraction within MCF7 cells was significantly increased. In addition, overexpression of PTEN decreased MCF7 cell tumorigenicity, which is consistent with reports on other tumors (31). Taken together, our data indicated that PTEN/PI3K/mTOR/STAT3 signaling forms a complex signaling network (Fig. 2) and maintains the cancer stem-like cell population within the whole cell population.
Fig. 2.
Schematic representation of PTEN/PI3K/mTOR/STAT3 signaling involved in breast cancer stem-like cell survival and proliferation. Arrows indicate activation, and bars indicate inhibition.
We noticed this study has some limitations. MCF7 cells have been propagated in vitro for many years and may not accurately reflect the behavior of human breast cancer in patients. In addition, MCF7 SP cells may be heterogeneous, and this study may actually underestimate the differences between breast cancer stem cells and bulk cancer cells. Further validation of cancer stem cells isolated from patient specimens is needed in future studies.
The promise of understanding cancer stem cell biology lies in the potential to provide new therapeutic approaches for cancer treatment. Current cancer drugs, which are developed extensively based on their activity to inhibit bulk replicating cancer cells, may not effectively inhibit cancer stem cells (14–16). It is conceivable that targeting cancer stem cells will be helpful in eradicating tumors more efficiently (14–16). One promising approach is to target cancer stem cell survival signaling pathways (14), where leukemia stem cell research has already made some progress (32). Through the use of a combined genomics and proteomics approach, which measured gene expression and phosphoprotein-based signaling, we identified signaling networks that appear to be important for breast cancer stem cell maintenance and viability. In particular, we found that PTEN/mTOR/STAT3 signaling is critical for SP cell survival and proliferation, which may provide targets for inhibiting cancer stem-like cells for improved treatment of cancer.
Materials and Methods
Hoechst 33342 Staining, Flow Cytometry Analysis, and Sorting of SP Cells.
Cells were washed with PBS, detached from the culture dish with trypsin and EDTA, pelleted by centrifugation, and resuspended in 37°C DMEM containing 2% FBS at 1 × 106 cells/ml. Cell staining was performed as described (11). The cells were incubated with Hoechst 33342 (Sigma, St. Louis, MO) at 5 μg/ml either alone or in combination with known ABC transporter inhibitors verapamil (50 μM, Sigma), reserpine (20 μM, Sigma), and FTC (10 μM, Sigma) for 90 min at 37°C. After staining, the cells were centrifuged and resuspended in HBSS (Invitrogen, Carlsbad, CA) containing 1 μg/ml propidium iodide and maintained at 4°C for flow cytometry analysis and sorting. Cell analysis and sorting were performed on a MoFlo cytometer (Dako Cytomation, Fort Collins, CO) equipped with a Coherent Enterprise II laser-emitting MLUV at 351- and blue 488-nm lines. The Hoechst 33342 emission was first split by using a 610-nm dichroic short-pass filter, and the red and the blue emissions were collected through 670/30- and 450/65-nm bandpass filters, respectively.
Cell Pellet Processing and Reverse-Phase Protein Arrays.
Cell pellets for the SP and non-SP populations from four independent experiments were collected and processed according to standard procedures (33). Arrays were prepared, blocked, stained with antibodies, and analyzed as described (33). Antibodies used in the experiments are listed in SI Table 7.
Statistical Analysis.
Statistical differences of two group data were compared by Student's t test (*, P < 0.05; **, P < 0.005). Statistical differences between non-SP and SP populations in protein array experiments were evaluated by a one-sample median test (or a hypothesis sign test for median) (34) by using SAS, Ver. 9.0 (SAS Institute, Cary, NC). Because the sample size of non-SP cells is four and the sample size of SP cells (pooled) is one, comparing the means of these two groups is not possible. However, it is reasonable to use the null-hypothesis test H0: Md = Msp, where Md and Msp are the median of the non-SP and the only value of the SP pooled groups, respectively. The P value is calculated by the binomial distribution table when sample size is four. The P value of the one-sided hypothesis test is 0.0625, and the P value of two-sided hypothesis test is 0.125. P < 0.0625 is considered statistically significant.
For more information, see SI Text.
Supplementary Material
Acknowledgments
We thank Xiaobing He for helpful discussions, Anne Jedlicka and Alan Scott for help with the microarray experiment, and Diane Griffin for encouragement. This work was supported by National Institutes of Health Grant AI44063, the Ho Ching Yang Memorial Faculty Fellowship in Cancer Prevention of the Johns Hopkins Bloomberg School of Public Health, the College of Science at George Mason University, and the Johns Hopkins Center for AIDS Research.
Abbreviations
- SP
side population
- ABC
ATP-binding cassette
- MDR
multidrug resistant
- shRNA
short-hairpin RNA
- RPA
reverse-phase signaling pathway protein array
- PI3K
phosphatidylinositol 3-kinase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702596104/DC1.
References
- 1.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien CA, Pollett A, Gallinger S, Dick JE. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 5.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 6.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Proc Natl Acad Sci USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 8.Kondo T, Setoguchi T, Taga T. Proc Natl Acad Sci USA. 2004;101:781–786. doi: 10.1073/pnas.0307618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 10.Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, Goodell MA, Brenner MK. Proc Natl Acad Sci USA. 2004;101:14228–14233. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou S, Schuetz JD, Bunting KD, Colapietro AM, Sampath J, Morris JJ, Lagutina I, Grosveld GC, Osawa M, Nakauchi H, Sorrentino BP. Nat Med. 2001;7:1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 13.Howe HL, Wingo PA, Thun MJ, Ries LA, Rosenberg HM, Feigal EG, Edwards BK. J Natl Cancer Inst. 2001;93:824–842. doi: 10.1093/jnci/93.11.824. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M, Rosen JM. Curr Opin Genet Dev. 2006;16:60–64. doi: 10.1016/j.gde.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Jones RJ, Matsui WH, Smith BD. J Natl Cancer Inst. 2004;96:583–585. doi: 10.1093/jnci/djh095. [DOI] [PubMed] [Google Scholar]
- 16.Reya T, Morrison SJ, Clarke MF, Weissman IL. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 17.Dean M, Fojo T, Bates S. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 18.Turkson J, Zhang S, Mora LB, Burns A, Sebti S, Jove R. J Biol Chem. 2005;280:32979–32988. doi: 10.1074/jbc.M502694200. [DOI] [PubMed] [Google Scholar]
- 19.Yokogami K, Wakisaka S, Avruch J, Reeves SA. Curr Biol. 2000;10:47–50. doi: 10.1016/s0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 21.Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 22.Furumoto Y, Brooks S, Olivera A, Takagi Y, Miyagishi M, Taira K, Casellas R, Beaven MA, Gilfillan AM, Rivera J. J Immunol. 2006;176:5167–5171. doi: 10.4049/jimmunol.176.9.5167. [DOI] [PubMed] [Google Scholar]
- 23.Abounader R, Reznik T, Colantuoni C, Martinez-Murillo F, Rosen EM, Laterra J. Oncogene. 2004;23:9173–9182. doi: 10.1038/sj.onc.1208146. [DOI] [PubMed] [Google Scholar]
- 24.Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, Nakauchi H, Taniguchi H. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 25.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 26.Paweletz CP, Charboneau L, Bichsel VE, Simone NL, Chen T, Gillespie JW, Emmert-Buck MR, Roth MJ, Petricoin IE, Liotta LA. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 27.Nishizuka S. Eur J Cancer. 2006;42:1273–1282. doi: 10.1016/j.ejca.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 28.Xu Q, Simpson SE, Scialla TJ, Bagg A, Carroll M. Blood. 2003;102:972–980. doi: 10.1182/blood-2002-11-3429. [DOI] [PubMed] [Google Scholar]
- 29.Ling X, Arlinghaus RB. Cancer Res. 2005;65:2532–2536. doi: 10.1158/0008-5472.CAN-04-2425. [DOI] [PubMed] [Google Scholar]
- 30.Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, Robbins PD, Gadiparthi S, Burke NA, Watkins SF, Grandis JR. Proc Natl Acad Sci USA. 2003;100:4138–4143. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheney IW, Johnson DE, Vaillancourt MT, Avanzini J, Morimoto A, Demers GW, Wills KN, Shabram PW, Bolen JB, Tavtigian SV, Bookstein R. Cancer Res. 1998;58:2331–2334. [PubMed] [Google Scholar]
- 32.Jordan CT, Guzman ML. Oncogene. 2004;23:7178–7187. doi: 10.1038/sj.onc.1207935. [DOI] [PubMed] [Google Scholar]
- 33.Wulfkuhle JD, Aquino JA, Calvert VS, Fishman DA, Coukos G, Liotta LA, Petricoin EF., III Proteomics. 2003;3:2085–2090. doi: 10.1002/pmic.200300591. [DOI] [PubMed] [Google Scholar]
- 34.Bowerman B, O'Connell R. Business Statistics in Practice. New York: McGraw–Hill; 2003. pp. 728–730. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.