Abstract
Epstein–Barr virus (EBV) was the first human virus found to encode microRNAs (miRNAs), but the function of these miRNAs has been obscure. Nasopharyngeal carcinoma (NPC) is associated with EBV infection, and the EBV-encoded LMP1 is believed to be a key factor in NPC development. However, detection of LMP1 protein in NPC is variable. Here, we report that EBV-encoded BART miRNAs target the 3′ UTR of the LMP1 gene and negatively regulate LMP1 protein expression. These miRNAs also modulate LMP1-induced NF-κB signaling and alleviate the cisplatin sensitivity of LMP1-expressing NPC cells. Consistent with a previous study on the NPC C666-1 cell line and C15 xenograft, we found abundant expression of BART miRNAs in NPC tissues. Furthermore, DNA sequencing revealed that the 3′ UTR of LMP1 is highly conserved in NPC-derived EBV isolates. The data provide insight into the discrepancy between LMP1 transcript and protein detection in NPC and highlight the role of the EBV miRNAs in regulating LMP1 downstream signaling to promote cancer development.
Keywords: Epstein–Barr virus, nasopharyngeal carcinoma
Epstein-Barr virus (EBV) is a ubiquitous herpesvirus that asymptomatically infects the majority of the population worldwide. EBV is also associated with the cancers Burkitt's lymphoma, Hodgkin's disease (HD), gastric carcinoma, and nasopharyngeal carcinoma (NPC) (1). In NPC, EBV expresses EBNA1, LMP1, and LMP2A, EBERs and the BamHI-A rightward transcripts (BARTs) (2). LMP1 can transform rodent fibroblasts and induce a wide range of phenotypic changes in epithelial cells and B cells in vitro (3–5). LMP1 activates the NFκB, JNK, JAK/STAT, p38/MAP, and Ras/MAPK pathways and alters cellular gene expression (6). LMP1 is thus a prime candidate for driving NPC development. Although immunohistochemical studies have found strong LMP1 expression in EBV-positive HD, detection of LMP1 protein in NPC tissues is variable (2, 6, 7). This raises questions about the contribution of LMP1 to NPC tumorigenesis.
The EBV BARTs were first identified in NPC as multispliced transcripts (8) and were later found in a wide range of EBV-associated cancers. BART expression is low in B lymphocytes and high in epithelial tissues (9–13), suggesting that the BARTs may be particularly important in epithelial malignancies. Although there are several small ORFs that can express proteins when cloned into heterologous vectors, definitive evidence for expression of BART proteins in vivo is limited (2, 6). The recent discovery of BART microRNAs (miRNA) has shed new light on the function of these transcripts. miRNAs are a class of ≈22 nucleotide noncoding RNAs that modulate gene expression by forming imperfect or perfect complementary duplexes with their target mRNAs, leading to translational inhibition or degradation of these mRNAs (14, 15). EBV was the first human virus reported to encode miRNAs (16). The EBV BARTs produce two clusters of miRNAs, 12 in Cluster 1 and 15 in Cluster 2. miR-BART2 is an individual miRNA [Fig. 1A and supporting information (SI) Table 3] (13, 16–18). Here, we report that BART Cluster 1 miRNAs target the LMP1 3′ UTR and negatively regulate LMP1 protein expression. BART miRNA regulation of LMP1 expression is reflected in modulation of LMP1-induced NF-κB activity, and suppression of cisplatin induced cytotoxicity in LMP1-expressing NPC cells.
Fig. 1.
Expression of EBV miRNAs in infected epithelial cells. (A) Diagram showing the EBV miRNA locations. Latency II promoters and genes expressed in NPC are black. Other latency promoters and genes are gray. BHRF1 is a lytic gene, and its promoter and coding region are indicated by the white pennant and box. (B) Northern blotting of BART Cluster 1 miRNAs in B95.8 B cells and the EBV-infected epithelial cell lines (1), C666-1 (2), CNE1-EBV (3), HK1-EBV (4), and AGS-BXI (5). U6 snRNA was used as a loading control. (C) Northern blotting of BART1-5p and BART2 miRNAs in NPC biopsies. B95–8 (positive control).
Results
Abundant Expression of BART miRNAs in EBV-Infected Cells.
The BART miRNAs have been characterized in B cell lines (13, 16, 17). We first compared the expression of BART Cluster 1 miRNAs in prototypic B95.8 B cells with four EBV-infected epithelial cell lines (Fig. 1B). In B95.8, the region that gives rise to a number of the BART miRNAs is deleted, and only BART 1-3p, 1-5p, 3-3p, 3-5p, and 4 were detected. In epithelial cells, the miRNA signal varied with more intense expression in C666-1 and AGS-BX1 than in CNE1-EBV and HK1-EBV. However, expression was detectable for all Cluster 1 miRNAs except BART 15. In all eight NPC biopsies examined, BART1-5p and BART2 miRNAs were also detected (Fig. 1C).
Interaction of BART Cluster 1 miRNAs with the LMP1 3′ UTR.
Using the default settings in the miRanda and MicroInspector programs, we determined that the 3′ UTR of LMP1 contains sequences with partial homology to BART1-5p, 3-5p, 4, 15, 16, 17-5p, and 17-3p (SI Fig. 7). To determine whether the LMP1 gene was a target of BART miRNAs, 10 predicted target sequences in the LMP1 3′ UTR (tL7, tL2, tL8, tL9, tL10, tL11, tL12, tL1, tL3, and tL6) of four miRNAs (BART3-5p, 16, 17-5p, and 1-5p) were examined (Fig. 2A). Four copies of each wild-type or mutant LMP1 target sequence (Fig. 2B) were cloned downstream from the stop codon in a luciferase vector. Each of the reporters was cotransfected with BART3-5p, 16, 17-5p, or 1-5p miRNA. miR-BART3-5p failed to suppress the luciferase activity of its target reporters (tL7 and tL2) (Fig. 2C), whereas the other three BART miRNAs significantly reduced the luciferase activity of their LMP1 target reporters. In particular, BART17-5p suppressed the luciferase activity of tL10 and tL12 by 77% and 90%, respectively, and miR-BART1-5p reduced the activity of tL6 by 94%. These findings suggest that BART Cluster 1 miRNAs functionally target the LMP1 3′ UTR.
Fig. 2.
BART Cluster 1 miRNAs target the LMP1 3′ UTR. (A) Schematic showing the location of predicted LMP1 target (tL) sites for BART 13-5p, 16, 17-5p, and 1-5p miRNAs. The LMP1 gene (open bar) is numbered according to GenBank accession no. X01995. Light shading, ORF. (B) Wild-type (wt) and mutant (mut) LMP1 3′ UTR target sequences of BART3-5p, 16, 17-5p, and 1-5p miRNAs that were cloned into luciferase reporters. (C) Luciferase activity of LMP1 target constructs in the presence of BART miRNAs. HeLa cells were transfected with luciferase reporters together with the indicated miRNA. The luciferase activity was normalized to β-gal activity. Fold change in luciferase activity of the wild-type LMP1 construct (black bars) was calculated relative to that of the mutated construct (white bars). Data shown are the mean ± SD from three separate experiments.
Differential Expression of LMP1 Protein and mRNA in EBV-Positive Epithelial Cells.
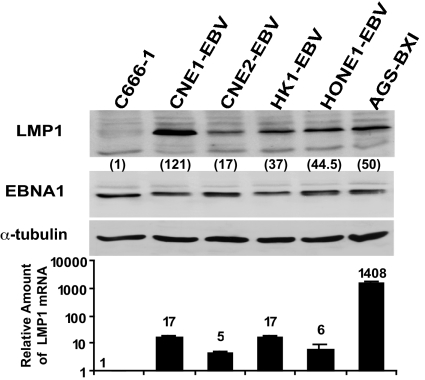
An examination of LMP1 protein and mRNA expression in EBV-infected epithelial cell lines (Fig. 3) found that LMP1 protein expression was not directly proportional to LMP1 mRNA levels. AGS-BX1 cells had 83-fold higher LMP1 mRNA levels but 2.4-fold less LMP1 protein than CNE1-EBV cells. Similarly, CNE1-EBV and HK1-EBV cells had the same LMP1 mRNA levels, but HK1-EBV cells expressed 3.2-fold less LMP1 protein. Immunohistochemical staining of LMP1 protein in two positive-control HD tissues (SI Fig. 8) showed strong LMP1 protein expression in both samples. Of the four NPC samples tested, LMP1 protein was not detected in NPC9 or NPC12 but was weakly positive in NPC11 and moderately positive in NPC10. A comparison of the relative LMP1 mRNA levels (calculated by using HD1 as the reference) and the relative levels of LMP1 protein (Table 1) found no direct correlation between the levels of mRNA and protein detected in these samples. For example, both NPC10 and NPC12 had a similar amount of LMP1 transcripts, but LMP1 protein was detected only in NPC10. These data suggest that LMP1 protein expression in EBV-positive cell lines and cancers is regulated not only at the transcriptional level but also by posttranscriptional mechanisms.
Fig. 3.
Comparison of LMP1 protein and mRNA expression in EBV-infected epithelial cells. (Upper) Western blotting of LMP1 protein in EBV-infected cells. Expression of EBNA1 and α-tubulin proteins formed internal controls. The relative LMP1 level was normalized to EBNA1. C666-1 was used as reference control and set at 1. (Lower) QRT-PCR of LMP1 mRNA in EBV-infected cells. Relative LMP1 level was normalized to EBNA1. C666-1 was set at 1.
Table 1.
Comparison of LMP1 protein and mRNA levels in EBV-positive NPC and HD specimens.
| Case | LMP1 protein (IHC) | Relative fold of LMP1 mRNA (QPCR) |
|---|---|---|
| NPC 9 | − | −5.3 |
| NPC 10 | ++ | 1.7 |
| NPC 11 | + | 2.2 |
| NPC 12 | − | 2.5 |
| HD 2 | +++ | 1,520.2 |
| HD 1 | +++ | 1 |
LMP1 mRNA level of HD1 was used as the reference control and set at 1. +, weak positive; ++, moderate positive; +++, strong positive; −, negative. IHC, immunohistochemistry; QPCR, quantitative PCR.
To compare BART miRNA and LMP1 expression in EBV-infected cells, Northern blot data from Fig. 1B was quantitated and compared with LMP1 protein and mRNA expression (from Fig. 3). The comparison for BART1-5p, 16, and 17 miRNAs (which have binding targets in the LMP1 3′ UTR as shown in Fig. 2) in the C666-1, CNE1-EBV, HK1-EBV, and AGS-BXI is presented in Table 2. CNE1-EBV and HK1-EBV expressed similar amounts of LMP1 mRNA. However, CNE1-EBV, which expressed a lower level of miRNAs than HK1-EBV, had higher LMP1 protein expression. Furthermore, AGS-BXI, which exhibited high miRNA expression, had lower LMP1 protein levels than did CNE1-EBV, which had a low BART miRNA abundance. Because of a limitation of tissue availability, a direct comparison of BART miRNA and LMP1 expression in NPC samples was not possible.
Table 2.
Comparison of BART miRNA, LMP1 mRNA, and protein expression in EBV-infected cells
| Expression | C666-1 | CNE1-EBV | HK1-EBV | AGS-BXI |
|---|---|---|---|---|
| BART1-5p miRNA | 1 | 0.1 | 0.19 | 0.8 |
| BART16 miRNA | 1 | 0.3 | 1.5 | 5 |
| BART17-5p miRNA | 1 | 0.2 | 0.1 | 2 |
| LMP1 mRNA | 1 | 17 | 17 | 1,048 |
| LMP1 protein | 1 | 121 | 37 | 50 |
BART Cluster 1 miRNAs Suppress LMP1 Protein Expression.
To substantiate the regulatory role of BART Cluster 1 miRNAs in LMP1 expression, a 2-kb DNA fragment covering the template for all Cluster 1 miRNAs was amplified from M-ABA and C666-1 EBV strains of NPC and cloned into a pCMV4 vector. These Cluster 1 vectors were confirmed to express all BART Cluster 1 miRNAs except BART15 (SI Fig. 9). The effect of Cluster 1 miRNAs on LMP1 expression was examined by using an LMP1 expression vector that included the LMP1 3′ UTR (LMP1 + 3′ UTR) and one lacking the 3′ UTR (LMP1 − 3′ UTR). The C666-1 Cluster 1 vector induced a dose-responsive reduction in LMP1 protein but not mRNA abundance in LMP1 + 3′ UTR transfected cells (Fig. 4A). However, no suppressive effect on LMP1 protein expression from the LMP1 − 3′ UTR vector was observed. The Cluster 1 vector also had no effect on EGFP control protein expression in any of the samples. These results indicate that the Cluster 1 miRNAs specifically suppress LMP1 protein expression at a posttranscriptional level by targeting the 3′ UTR of LMP1. The dose-responsive reduction of LMP1 protein expression from the LMP + 3′ UTR plasmid was also observed after cotransfection with a MABA-Cluster 1 vector or an individual BART1-5p miRNA (Fig. 4B). However, when a Cluster 2 vector or a synthetic BART2 miRNA was cotransfected, no suppression was seen. In a further test using individual BART16, 17-5p, or 1-5p miRNAs, LMP1 protein expression from the LMP + 3′ UTR vector was reduced, whereas expression from LMP1 − 3′ UTR vector was not affected (Fig. 4C). To further support the specificity of the Cluster 1 miRNAs in suppressing LMP1 protein expression, the Cluster 1 vector was cotransfected with vectors expressing EBV Zta or Rta or cellular APOBEC3G or IRF7 proteins. No significant reduction in the expression of any of these proteins was observed (SI Fig. 10).
Fig. 4.
Suppression of LMP1 protein expression by BART Cluster 1 miRNAs (A Upper) Western blotting of LMP1 and control EGFP in HeLa cells transfected with EGFP, LMP1, and Cluster 1 plasmids as indicated. LMP1 level was normalized to EGFP and was calculated relative to the vector control, which is set at 1. (Lower) LMP1 mRNA in the same samples assayed by QRT-PCR. LMP1 mRNA is normalized to EBNA1 and is shown relative to the vector control (set at 1). (B) HeLa cells were cotransfected with EGFP vector, LMP1 + 3′ UTR plasmid, and MABA-Cluster 1, Cluster 2 vector (Upper), BART1-5p, or BART2 synthetic miRNA (Lower). (C) The LMP1 expression vectors were transfected into HeLa cells together with the EGFP vector and miRNAs as indicated. (D Left) CNE1-EBV cells transfected with miRNAs as indicated. (Center) CNE1-EBV cells transfected with anti-miRNA oligos as indicated. (Right) AGS-BXI cells transfected with Cluster 1 vectors as indicated. EBNA1 and α-tubulin protein served as internal controls. LMP1 expression was normalized to EBNA1. (Lower) QRT-PCR of LMP1 mRNA in the same samples. LMP1 mRNA expression was normalized to EBNA1.
Additionally, transfection of CNE1-EBV cells with individual BART16, 17-5p, or 1-5p miRNAs reduced endogenous LMP1 protein expression 27–72% (Fig. 4D Left). In contrast, transfection of antisense oligonucleotides for miR-BART16, 17-5p or 1-5p into CNE1-EBV cells enhanced LMP1 protein expression 1.7- to 3.2-fold (Fig. 4D Center). Increased expression of the entire set of Cluster 1 miRNAs in AGS-BXI by transfection of MABA or C666-1 Cluster 1 plasmids reduced LMP1 protein levels by 56% and 51% (Fig. 4D Right). No reduction of EBV-encoded EBNA1 or cellular α-tubulin proteins was seen in any of the samples examined. There was also no significant change in LMP1 mRNA abundance.
BART Cluster 1 miRNAs Reduce the Sensitivity of LMP1-Expressing Cells to Cisplatin.
LMP1 promotes cellular transformation. However, overexpression of LMP1 can be growth-inhibitory and potentiate apoptosis induced by stresses or treatment with chemotherapeutic agents such as cisplatin (19, 20), suggesting that LMP1 function is dose-sensitive. The potential role of BART Cluster 1 miRNAs in modulating the toxic effect of LMP1 was examined in the context of cisplatin treatment. Consistent with previous data (20), a colony-forming assay showed that LMP1 expression in CNE2 cells enhanced cisplatin-induced toxicity (Fig. 5A). The IC50 value decreased from 69 ± 3 ng/ml (control) to 46 ± 10 ng/ml (LMP1). However, coexpression of the Cluster 1 vector with LMP1 protected the cells from cisplatin-induced toxicity and shifted the IC50 value to 84 ± 17 ng/ml (Cluster 1 + LMP1). PARP protein cleavage also increased upon transfection of LMP1 (Fig. 5B). The same amount of transfected LMP1 did not increase PARP cleavage in the presence of Cluster 1 miRNAs, in part because of a reduction in LMP1 protein expression. In a TUNEL assay (Fig. 5C), introduction of Cluster 1 miRNAs reduced apoptosis, particularly at the highest dose of cotransfected LMP1.
Fig. 5.
Cluster 1 miRNAs attenuate LMP1-induced cytotoxicity to cisplatin. (A) Colony-forming assay. CNE2 cells cotransfected with control vector, LMP1 alone, or plus the Cluster 1 expression vector were trypsinized and seeded into six-well plates. Cells were treated with cisplatin, and colonies consisting of >50 cells were scored on day 8. Data shown are the mean ± SD from three separate experiments. (B) Western blotting of PARP protein cleavage in CNE2 cells cotransfected with LMP1 together with pCMV4 or C666-1-Cluster 1 plasmid and treated with 0.5 μg/ml cisplatin. The relative PARP cleavage was calculated after normalizing to α-tubulin protein. The LMP1-null-transfected samples were set at 1. LMP1 and EGFP were included as internal controls. (C) CNE2 cells were transfected with LMP1 vector and either pCMV4 or C666-1-Cluster 1 constructs. Apoptotic cells were measured by TUNEL assay after 1 day of cisplatin treatment. Data shown are the mean ± SD from three experiments.
BART Cluster 1 miRNAs Modulate Downstream LMP1 Activation of NF-κB.
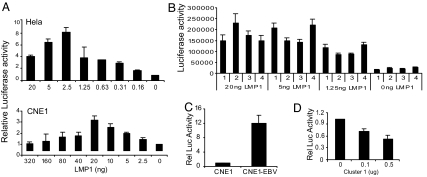
NF-κB signaling is the major pathway activated by LMP1 and is a key contributor to LMP1-induced transformation (3). An examination of LMP1 activation of NF-κB in HeLa and CNE1 cells (Fig. 6A) revealed that increasing the amount of LMP1 increased NF-κB activity up to a certain point, after which additional LMP1 led to a gradual reduction in NF-κB activity. Interestingly, the threshold amounts of LMP1 that resulted in peak NF-κB activity were different, with 2.5 ng of LMP1 in HeLa and 20 ng in CNE1 producing maximal activation. To address the possibility that the reduction of NF-κB activity with increasing amounts of LMP1 might reflect LMP1-induced cell death or growth inhibition, HeLa and CNE1 cells were cotransfected with a luciferase reporter together with vector control or 250 ng of the LMP1 expression vector. This amount of LMP1 was 50- and 6.25-fold higher than the amount at which NF-κB activity declined in HeLa and CNE1 cells, respectively. At 2 days after transfection, the luciferase activity in LMP1-transfected cells was 2.0- to 2.2-fold higher than in the vector controls (SI Fig. 11), indicating that the reduction in LMP1-induced NF-κB activity was occurring in the absence of growth inhibition.
Fig. 6.
Modulation of LMP1-induced NF-κB activity by Cluster 1 miRNAs. (A) NF-κB luciferase reporter activity was measured in HeLa (Upper) or CNE1 (Lower) cells cotransfected with increasing amounts of LMP1 vector plus an SV40-β-gal control. Luciferase activity was normalized to β-gal activity and was plotted relative to that of reporter alone (set at 1). (B) HeLa cells were cotransfected with the NF-κB luciferase reporter, pCMV4 control (1), MABA-Cluster 1 (2), C666-1-Cluster 1 (3), or Cluster 2 vector (4) together with different amounts of LMP1 as indicated. Data represent the mean of normalized luciferase/β-gal activity ± SD from three independent experiments. (C) NF-κB activity was measured in CNE1 and CNE1-EBV cells transfected with an NFκB luciferase reporter vector. NF-κB activity is plotted relative to that in CNE1 cells (set at 1). (D) CNE1-EBV cells were cotransfected with the NF-κB luciferase reporter and the Cluster 1 miRNA plasmid. NF-κB activity is plotted relative to that of the reporter alone (set at 1).
To examine whether Cluster 1 miRNAs can modulate LMP1-induced NF-κB signaling, HeLa cells were cotransfected with an NF-κB-luciferase reporter and increasing amounts of an LMP1 expression vector together with Cluster 1 or Cluster 2 miRNA plasmids (Fig. 6B). Neither Cluster 1 nor Cluster 2 miRNAs affected NF-κB activity in the absence of LMP1 (0 ng LMP1). The NF-κB activation induced by cotransfection of 1.25 or 5 ng of LMP1 was down-regulated by MABA and C666-1 Cluster 1 miRNAs and was not affected by Cluster 2 miRNAs. However, in the presence of 20 ng of LMP1, the Cluster 1 miRNAs no longer suppressed but, instead, increased the NF-κB reporter activation. These data indicate that Cluster 1 miRNAs can modulate LMP1-induced NF-κB activity. A comparison of NF-κB activity in CNE1 and CNE1-EBV cells found a 12-fold increase in the EBV-infected CNE1 cells (Fig. 6C). Introduction of Cluster 1 miRNAs into CNE1-EBV cells resulted in a dose-dependent suppression of NF-κB activity (Fig. 6D). Thus, the Cluster 1 miRNAs not only modulate LMP1 protein expression but also, in turn, modify LMP1-induced NF-κB signaling in EBV-infected cells.
Sequence Analysis of the LMP1 3′ UTR from Different EBV Isolates.
A comparison of the sequence of the 1,219-bp LMP1 3′ UTR for three EBV strains from NPC: GD1 (AY961628), 2117 (EF110909), and C666-1 (EF103558) with the prototypic B95.8, revealed an average of three to six nucleotide changes (SI Fig. 12). In EBV isolates from 10 primary NPCs examined (SI Fig. 13), all had three common nucleotide changes, and these were the only nucleotide changes in seven of the isolates. Only the predicted target sequences for miR-BART3-5p are affected by any of the changes. However, miR-BART3-5p was not functional in our assays and may not target the LMP1 3′ UTR. Overall, the LMP1 3′ UTR is conserved in NPC isolates, and this contrasts with the LMP1 coding region where frequent mutations have been reported (21).
Discussion
In this study, we showed that the EBV-encoded BART Cluster 1 miRNAs target the 3′ UTR of the LMP1 gene and modulate LMP1 protein expression. A lack of correlation between LMP1 mRNA and protein expression in the different EBV-infected epithelial cells and NPC tissues was also noted. We ascribe this to the activity of the miRNAs.
The current data suggest that regulation of LMP1 protein in NPC can occur at both transcriptional and posttranscriptional levels. Methylation of the LMP1 5′ regulatory region has been noted in NPCs with undetectable LMP1 expression (22, 23). The expression of lytic LMP1 has been proposed as a posttranslational regulatory mechanism (24). However, the promoter that drives the lytic form of LMP1 protein is consistently absent from EBV strains isolated from NPC. In line with a previous study (13), we found that BART miRNAs are abundantly expressed in EBV-infected cell lines and NPC tissues (Fig. 1). Sequence analysis revealed that the LMP1 3′ UTR that is targeted by the Cluster 1 miRNAs is conserved in EBV strains of NPC. An inverse correlation between the BART miRNA expression and LMP1 protein expression was also observed. The BART Cluster 1 miRNAs thus provide a means of posttranscriptional negative regulation of LMP1 in NPC.
Although LMP1 is capable of inducing cell growth and transformation, elevated expression of LMP1 can result in growth inhibition (25) and sensitization to apoptosis after stimuli such as starvation, treatment with TNFα, Fas, or chemotherapeutic agents (19, 20, 26, 27). In addition, a similar level of LMP1 expression driven from the same retroviral vector can induce paradoxical effects in different cell lines. In the immortalized NP69 nasopharyngeal epithelial cell line (28), LMP1 promotes cell growth and resistance to serum deprivation-induced apoptosis, whereas LMP1 induces growth inhibition and increased sensitivity to cisplatin-induced cell death in the CNE2 NPC cell line (20). These observations reflect the complexity of cellular signaling induced by LMP1 and indicate that the overall outcome represents a combination of the intrinsic properties of the cell and the LMP1 protein level. In this study, we demonstrated that Cluster 1 miRNAs attenuate the apoptotic effect of cisplatin in LMP1-expressing CNE2 cells. Negative regulation of LMP1 expression may favor EBV-associated cancer development by balancing the growth advantages and disadvantages of LMP1.
LMP1 is believed to be important to NPC development, but LMP1 protein expression in NPC is variable. LMP1 protein is detected by immunoblotting or immunohistochemistry in 20–68% of NPC cases (29). In contrast, with the exception of a recent report (30), RT-PCR analyses have found LMP1 mRNA expression in the majority of NPC samples (29). A discrepancy between LMP1 mRNA and protein levels was also seen in this study (Fig. 3). The lack of LMP1 protein expression in some NPC samples may not accurately reflect the contribution of LMP1 to the pathology of NPC. In the transgenic mouse model, LMP1 expression led to epithelial hyperplasia, and yet LMP1 protein could not be readily detected with standard extraction methods (31). Here, we found that very small amounts of LMP1 that may be difficult to detect with currently available reagents, are sufficient to activate NF-κB. NF-κB is activated by LMP1 through CTAR1 and CTAR2 domains that can engage TRAFs, TRADD, and RIP. LMP1 induces both canonical IKK-dependent and noncanonical NIK-dependent NF-κB pathways (32). The NF-κB transcription factors influence proliferation, apoptosis, oncogenesis, and inflammation (33). We found that low amounts of LMP1 stimulated NF-κB activity in epithelial cells, whereas an increased amount of LMP1, one that was still associated with cell-growth promotion, reduced NF-κB activity. Integration of signaling from the two CTAR domains and integration of different NFκ-B signal components and pathways influence the specificity, strength, and duration of signal activity and may be affected by LMP1 protein levels. The ability of BART miRNAs to modulate NF-κB activity through regulation of LMP1 protein levels may be an important component of their functioning. Our finding that BART Cluster 1 miRNAs regulate LMP1 expression in NPC provides insight into the discrepancy between LMP1 mRNA and protein abundance in NPC and highlights the importance of tightly controlled LMP1 protein expression for EBV-associated pathogenesis in epithelial cells. The data raise the possibility that negating the effects of the BART miRNAs may provide a therapeutic strategy for NPC treatment.
Materials and Methods
DNA Constructs and miRNA.
The MABA- and C666-1-Cluster 1 and the Cluster 2 miRNA plasmids have 1.8- and 3.6-kb DNA fragment inserts, respectively, generated by PCR and cloned into the pCMV4 vector. The LMP1 + 3′ UTR and LMP1 − 3′ UTR fragments were cloned into pCDNA3.1 (Invitrogen, Carlsbad, CA). The LMP1 luciferase reporters were cloned with four copies of LMP1 target sequence downstream from the stop codon of pGL3 control vector (Promega, Madison, WI). The NF-κB luciferase reporter is from Stratagene (La Jolla, CA). The nomenclature of the BART miRNAs is from the miRBase sequence database (see SI Methods for details).
Cell Lines, Tumors, and Transfection.
All NPC cell lines were cultured in RPMI medium 1640. AGS-BXI gastric cancer cells were grown in F-12 medium. C666-1 is an EBV-harboring NPC cell line. All other EBV-positive epithelial cell lines were stably infected with recombinant Akata EBV. Primary NPC tumors were obtained from the pathology bank of the Department of Anatomical and Cellular Pathology at the Chinese University of Hong Kong. DNA plasmids and miRNA transfections were performed by using Lipofectamine 2000 (Invitrogen) (see SI Methods for details).
Western Blotting.
Western blotting was performed as described (28). The primary antibodies used were LMP1 (DAKO, Glostrup, Denmark), α-tubulin (Sigma, St. Louis, MO), EGFP (Clontech, Mountain View, CA), and PARP (Cell Signaling Technology, Beverly, MA).
Luciferase Reporter Assay.
Cells (1 × 105) grown in 24-well plates were cotransfected with 10 pmol of miRNA and 25 ng of luciferase reporter constructs. SV40-β-gal vector (25 ng) was transfected as an internal control to normalize the transfection efficiency. Two days after transfection, cells were lysed in reporter lysis buffer (Promega) and then assayed for luciferase and β-gal activities.
Northern Blotting.
Total RNA was purified by using TRIzol reagent (Invitrogen). Total RNA from each sample was electrophoresed on 15% SDS/PAGE gels and electroblotted onto nylon membrane. Northern blotting was performed by using PerfectHyb Plus hybridization buffer (Sigma). Hybridization signals were visualized by either autoradiography or PhosphorImaging (Molecular Dynamics, Sunnyvale, CA). For probes used, see SI Methods.
Quantitative RT-PCR (QRT-PCR).
All QRT-PCR products were amplified by using SYBR green PCR Master Mix kit (Applied Biosystems, Foster City, CA). The values for the relative quantification were calculated by the ΔΔCT method. Data are shown with the value of the reference sample set at 1. For quantification of LMP1 mRNA in EBV-infected cells, Qp-EBNA1 was used as a control. Primers for LMP1 and Qp-EBNA1 have been described (34, 35).
TUNEL Assay.
The TUNEL assay was performed by using an In Situ Cell Death Fluorescein Detection kit (Roche, Indianapolis, IN).
EBER in Situ Hybridization (ISH) and Immunohistochemical (IHC) Staining of LMP1.
ISH and IHC were performed by standard methods. See SI Methods for details.
Supplementary Material
Acknowledgments
This work was supported by a Croucher Foundation Fellowship (to A.K.F.L.) and U.S. Public Health Service Grants CA30356 and CA42245 (to S.D.H.). K.W.L. and K.F.T. were supported by Li Ka Shing Institute of Health Science and Kadoorie Charitable Foundations.
Abbreviations
- BART
BamHI-A rightward transcript
- HD
Hodgkin's disease
- miRNA
microRNA
- NPC
nasopharyngeal carcinoma.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequences reported in this paper have been deposited in the GenBank database (accession nos. EF110909 and EF103558.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702896104/DC1.
References
- 1.Rickinson AB, Kieff E. In: Fields Virology. Knipe DM, Howley PM, Griffin DE, editors. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2575–2625. [Google Scholar]
- 2.Raab-Traub N. Semin Cancer Biol. 2002;12:431–441. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- 3.Izumi KM, Kieff ED. Proc Natl Acad Sci USA. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kieff E, Rickinson AB. In: Fields Virology. Knipe DM, Howley PM, Griffin DE, editors. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2511–2573. [Google Scholar]
- 5.Young LS, Murray PG. Oncogene. 2003;22:5108–5121. doi: 10.1038/sj.onc.1206556. [DOI] [PubMed] [Google Scholar]
- 6.Young LS, Rickinson AB. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 7.Deacon EM, Pallesen G, Niedobitek G, Crocker J, Brooks L, Rickinson AB, Young LS. J Exp Med. 1993;177:339–349. doi: 10.1084/jem.177.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith P. Semin Cancer Biol. 2001;11:469–476. doi: 10.1006/scbi.2001.0414. [DOI] [PubMed] [Google Scholar]
- 9.Karran L, Gao Y, Smith PR, Griffin BE. Proc Natl Acad Sci USA. 1992;89:8058–8062. doi: 10.1073/pnas.89.17.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilligan KJ, Rajadurai P, Lin JC, Busson P, Abdel-Hamid M, Prasad U, Tursz T, Raab-Traub N. J Virol. 1991;65:6252–6259. doi: 10.1128/jvi.65.11.6252-6259.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith PR, de Jesus O, Turner D, Hollyoake M, Karstegl CE, Griffin BE, Karran L, Wang Y, Hayward SD, Farrell PJ. J Virol. 2000;74:3082–3092. doi: 10.1128/jvi.74.7.3082-3092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen HL, Lung MM, Sham JS, Choy DT, Griffin BE, Ng MH. Virology. 1992;191:193–201. doi: 10.1016/0042-6822(92)90181-n. [DOI] [PubMed] [Google Scholar]
- 13.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Cullen BR. Nat Genet. 2006;38(Suppl):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 16.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 17.Grundhoff A, Sullivan CS, Ganem D. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu JJ, Chen JY, Hsu TY, Yu WC, Su IJ, Yang CS. J Gen Virol. 1996;77(Pt 8):1883–1892. doi: 10.1099/0022-1317-77-8-1883. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Wang X, Lo AK, Wong YC, Cheung AL, Tsao SW. J Med Virol. 2002;66:63–69. doi: 10.1002/jmv.2112. [DOI] [PubMed] [Google Scholar]
- 21.Edwards RH, Sitki-Green D, Moore DT, Raab-Traub N. J Virol. 2004;78:868–881. doi: 10.1128/JVI.78.2.868-881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Minarovits J. Adv Cancer Res. 2003;89:133–156. doi: 10.1016/s0065-230x(03)01004-2. [DOI] [PubMed] [Google Scholar]
- 23.Hu LF, Minarovits J, Cao SL, Contreras-Salazar B, Rymo L, Falk K, Klein G, Ernberg I. J Virol. 1991;65:1558–1567. doi: 10.1128/jvi.65.3.1558-1567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandya J, Walling DM. J Virol. 2006;80:8038–8046. doi: 10.1128/JVI.00180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eliopoulos AG, Dawson CW, Mosialos G, Floettmann JE, Rowe M, Armitage RJ, Dawson J, Zapata JM, Kerr DJ, Wakelam MJ, et al. Oncogene. 1996;13:2243–2254. [PubMed] [Google Scholar]
- 26.Liu MT, Chen YR, Chen SC, Hu CY, Lin CS, Chang YT, Wang WB, Chen JY. Oncogene. 2004;23:2531–2539. doi: 10.1038/sj.onc.1207375. [DOI] [PubMed] [Google Scholar]
- 27.Kawanishi M. Virology. 2000;270:258–266. doi: 10.1006/viro.2000.0296. [DOI] [PubMed] [Google Scholar]
- 28.Lo AK, Liu Y, Wang XH, Huang DP, Yuen PW, Wong YC, Tsao GS. Lab Invest. 2003;83:697–709. doi: 10.1097/01.lab.0000067480.44925.10. [DOI] [PubMed] [Google Scholar]
- 29.Tsao SW, Tramoutanis G, Dawson CW, Lo AK, Huang DP. Semin Cancer Biol. 2002;12:473–487. doi: 10.1016/s1044579x02000901. [DOI] [PubMed] [Google Scholar]
- 30.Bell AI, Groves K, Kelly GL, Croom-Carter D, Hui E, Chan AT, Rickinson AB. J Gen Virol. 2006;87:2885–2890. doi: 10.1099/vir.0.81906-0. [DOI] [PubMed] [Google Scholar]
- 31.Stevenson D, Charalambous C, Wilson JB. Cancer Res. 2005;65:8826–8835. doi: 10.1158/0008-5472.CAN-05-0591. [DOI] [PubMed] [Google Scholar]
- 32.Luftig M, Yasui T, Soni V, Kang MS, Jacobson N, Cahir-McFarland E, Seed B, Kieff E. Proc Natl Acad Sci USA. 2004;101:141–146. doi: 10.1073/pnas.2237183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rayet B, Gelinas C. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 34.Goormachtigh G, Ouk TS, Mougel A, Tranchand-Bunel D, Masy E, Le Clorennec C, Feuillard J, Bornkamm GW, Auriault C, Manet E, et al. J Virol. 2006;80:7382–7393. doi: 10.1128/JVI.02052-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Hutt-Fletcher L, Cao L, Hayward SD. J Virol. 2003;77:4139–4148. doi: 10.1128/JVI.77.7.4139-4148.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.