Abstract
Mucins are proteins that cover and protect epithelial cells and are characterized by domains rich in proline, threonine, and serine that are heavily glycosylated (PTS or mucin domains). Because of their sequence polymorphism, these domains cannot be used for evolutionary analysis. Instead, we have made use of the von Willebrand D (VWD) and SEA domains, typical for mucins. A number of animal genomes were examined for these domains to identify mucin homologues, and domains of the resulting proteins were used in phylogenetic studies. The frog Xenopus tropicalis stands out because the number of gel-forming mucins has markedly increased to at least 25 as compared with 5 for higher animals. Furthermore, the frog Muc2 homologues contain unique PTS domains where cysteines are abundant. This animal also has a unique family of secreted mucin-like proteins with alternating PTS and SEA domains, a type of protein also identified in the fishes. The evolution of the Muc4 mucin seems to have occurred by recruitment of a PTS domain to AMOP, NIDO, and VWD domains from a sushi domain-containing family of proteins present in lower animals, and Xenopus is the most deeply branching animal where a protein similar to the mammalian Muc4 was identified. All transmembrane mucins seem to have appeared in the vertebrate lineage, and the MUC1 mucin is restricted to mammals. In contrast, proteins with properties of the gel-forming mucins were identified also in the starlet sea anemone Nematostella vectensis, demonstrating an early origin of this group of mucins.
Keywords: bioinformatics, von Willebrand domain, SEA domain, protein evolution, mucus
All mucosal membranes of the body are covered by mucus, largely made up of the family of large glycoproteins called mucins. Mucins may be defined as having one long mucin domain rich in proline, threonine, and serine (PTS domain), which is heavily glycosylated through GalNAc O-linkage to the serine and threonine residues. With this definition of mucins, 14 proteins have been assigned to the MUC gene family according to the HUGO nomenclature (www.genenames.org), although this family contains proteins that differ considerably. Other proteins with shorter mucin domains, like IgA and several receptors, are not called mucins. The mucin domains often contain tandemly repeated sequences and vary to a great extent in sequence and length depending on allele, species, and individual mucin (1, 2). The mucins have been further classified as membrane-bound or secreted. In humans there are eight membrane-bound (MUC1, MUC3, MUC4, MUC12, MUC13, MUC16, MUC17, and MUC20) and five secreted gel-forming mucins (MUC2, MUC5B, MUC5AC, MUC6, and MUC19 (1, 2).
All of the transmembrane mucins are type 1 proteins with one membrane spanning domain but are also characterized by other protein domains. The human MUC1, MUC3, MUC12, MUC13, MUC16, and MUC17 mucins contain SEA domains, and the MUC4 mucin NIDO, AMOP, and VWD domains. These mucins reach far out from the cell, but their functions are so far unknown. However, both their cytoplasmic tails as well as the structure of the SEA domain suggest that they are involved in various signaling events as best studied for the MUC1 mucin (2, 3). Typically, these mucins are found apically in polarized epithelia, but are distributed over the whole cell in cancer. Several of these mucins are also involved in the progression of cancer and are often linked to poor prognosis (2).
The human gel-forming mucins contain VWD and C-terminal cysteine-knot domains that are responsible for the oligomerization of these molecules. The cysteine-knot is responsible for dimerization in the endoplasmic reticulum, and, in the case of porcine submaxillary mucin (PSM) and MUC2, the third von Willebrand D (VWD) domain in its N-terminal part is responsible for trimerization in the late secretory pathway (1, 4). The gel-forming mucins are instrumental in the protection of the underlying epithelia and in trapping and elimination of bacteria. Although our understanding of these mucins at the molecular level is still rudimentary, they are clearly involved in the pathogenesis of several diseases. Inflammation of the large intestine and the inflammatory bowel disease ulcerative colitis is observed in mice that are deficient in the mucin MUC2 (5), suggesting that the mucins are important for intestinal handling of bacteria. Overproduction of mucins in the lungs is a key element of both chronic obstructive lung disease and cystic fibrosis. In insects, the peritrophic matrix of the gut has a protective role similar to that of the mucus of higher animals, but the insect matrix is made up of chitin and a number of chitin-binding proteins, of which some also contain mucin domains (6).
Our understanding of mucins has been hampered by experimental difficulties because of their large size, oligomerization, and extensive glycosylation. To facilitate studies of mucins, we reasoned that a better knowledge of the evolution and distribution of mucins should be of help. Along these lines, we have previously exploited the typical mucin domains as well as PTS domains to predict mucin proteins computationally in fish and chicken (7, 8). The rapid increase in the number of sequenced metazoan genomes has now made it possible to extend such studies in an attempt to further understand the molecular biology of mucins. Therefore, we have made use of available genomes to systematically examine phylogeny and evolution of mucins containing the VWD and SEA domains.
Results
Mucins are characterized by PTS (or mucin) domains, regions rich in the amino acids serine, threonine, and proline. PTS domains tend to be poorly conserved in sequence and are therefore not useful for an examination of the evolutionary relationships between mucins. On the other hand, most mucins are characterized by specific domains, such as the VWD or SEA domains that can be used for such studies. Therefore, we first identified all possible proteins with VWD and SEA domains. Typically, these proteins were identified by using the Pfam models for VWD and SEA (9). In some cases, genomes are poorly characterized, and we used Genewise to scan genome sequences with the Pfam VWD/SEA domain models. The identified sequences were analyzed for neighboring PTS domains by using PTSPred (7), and their domain structure was analyzed in general, including Pfam domains, signal sequences, and transmembrane domains (TMs). We also analyzed the exon structure of all potential mucins to understand gene structure and to predict their amino acid sequences. We analyzed a range of metazoa, including mammals, chicken (Gallus gallus), frog (Xenopus tropicalis), fishes (Danio rerio, Takifugu rubripes, and Tetraodon nigroviridis), the sea squirt Ciona intestinalis, the lancelet Branchiostoma floridae, the sea urchin Strongylocentrotus purpuratus, the fruit fly Drosophila melanogaster, the worm Caenorhabditis elegans, and the starlet sea anemone Nematostella vectensis. For information on all identified mucins and related proteins, including chromosome localization, EST support of predicted genes, PTS repeat information, and supplementary phylogenetic trees, the reader is referred to the supplementary material and our website: www.medkem.gu.se/mucinbiology/databases.
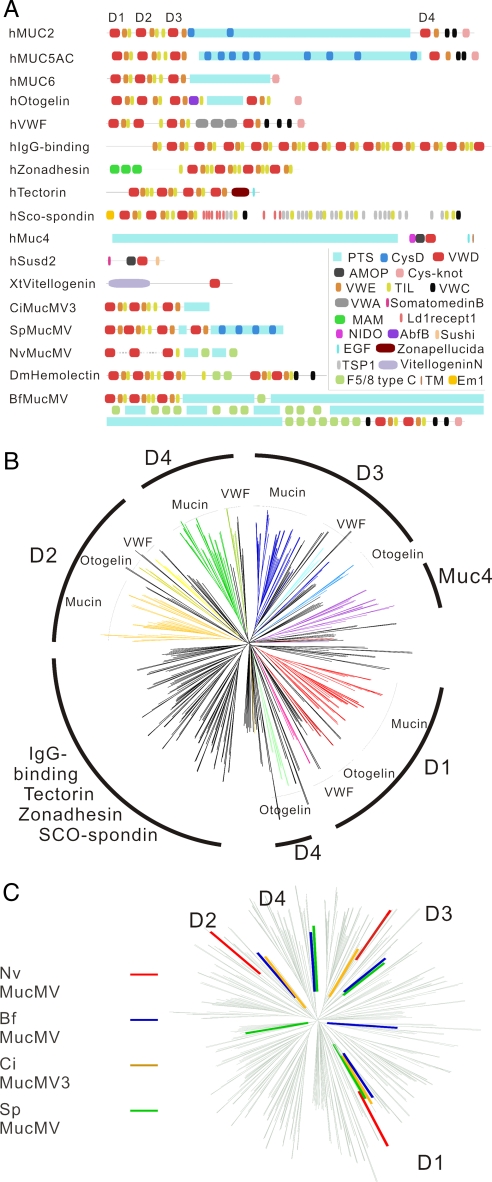
Many VWD Domain Proteins Contain a Conserved Arrangement of VWD, VWE, and TIL Domains.
A number of proteins in addition to mucins are known to contain VWD domains, such as the von Willebrand factor, otogelin, SCO-spondin, tectorin, zonadhesin, vitellogenin, IgG-binding protein, hemolectin, and Susd2. VWD domains are often tandemly arranged with three domains, sometimes with a fourth domain as shown for mucins and some other proteins in Fig. 1A. We refer to these domains as D1, D2, D3, and D4, based on their position in the protein (Fig. 1A). The VWD domains often occur together with a Pfam domain referred to as DUF1787 and TIL (trypsin inhibitor-like) domains. Here, we will refer to the DUF1787 domain as “VWE” (von Willebrand E domain). This is a domain rich in cysteines that seems be present only in the context of a VWD domain. In addition to the VWD, VWE, and TIL domains, the different VWD families are characterized by additional domains, such as VWA in the von Willebrand factor, the PTS domain in mucins, F5/8 type C (discoidin) domain in hemolectin, and the AMOP and NIDO domains in Muc4 (Fig. 1A).
Fig. 1.
Proteins with VWD domains. (A) Domain structures of selected proteins containing the VWD domain, including proteins showed as colored branches in C. Organisms represented are human (h), X. tropicalis (Xt), C. intestinalis (Ci), S. purpuratus (Sp), N. vectensis (Nv), D. melanogaster (Dm), and B. floridae (Bf). (B) Phylogenetic tree of VWD domains. Four hundred thirty-one VWD domains from 147 proteins were aligned by using ClustalW, and shown in the figure is the neighbor-joining tree derived by the same program. Branches with the D1, D2, D3, and D4 domains (A) corresponding to the gel-forming mucins otogelin and VWF are indicated as well as Muc4 and a branch containing the group of proteins (IgG-binding, tectorin, zonadhesin, and SCO-spondin) that do not group with the D1–D4 domains. The D1–D4 domains are shown with different colors. A rectangular version of this tree with more details on the individual branches as well as bootstrapping data is shown at www.medkem.gu.se/mucinbiology/databases. (C) Tree as in B but with VWDs of proteins of N. vectensis (red), S. purpuratus (green), B. floridae (blue), and C. intestinalis (yellow). All of the VWD domains of these proteins clearly map to the D1–D4 clusters shown in B.
Gel-Forming Mucins, Otogelin, von Willebrand Factor Are Evolutionary Related.
We have identified a total of 431 VWD domains in 147 proteins from a range of animals as listed above. These were aligned with ClustalW and analyzed with neighbor-joining and parsimony methods [Fig. 1B, supporting information (SI) Fig. 5, and www.medkem.gu.se/mucinbiology/databases] The VWE and TIL domains were also used in phylogenetic studies and gave rise to trees very similar in topology to that in Fig. 1B.
The phylogenetic analysis helps to clarify the relationship between different VWD domains, aids in the classification of mucins, and is useful in an examination of the evolution of mucins as related to other VWD-containing proteins. Most importantly, we are able to identify homologues to the gel-forming mucins in the lower animals.
The phylogenetic analysis (Fig. 1B) shows that the D1, D2, D3, and D4 domains of vertebrate gel-forming mucins, otogelin, and VWF are related and that these proteins are phylogenetically distinct from the other proteins with multiple VWD domains, i.e., tectorin, zonadhesin, SCO-spondin, and IgG-binding proteins. This relationship is even more obvious when only vertebrate sequences are included in the analysis (data not shown).
Gel-Forming Mucins in Nonvertebrate Chordates.
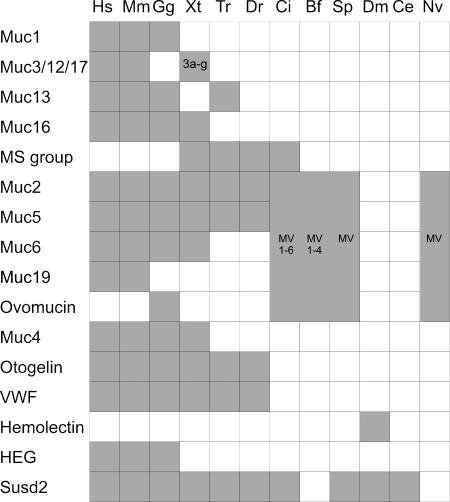
We identified mucins and mucin-related proteins in a variety of vertebrates as well as lower animals such as C. intestinalis, B. floridae, and S. purpuratus (Fig. 2). Gel-forming mucins are characterized by three VWD domains, followed by a PTS region, and proteins with these characteristics were identified in all vertebrates including teleosts. X. tropicalis contains a large number of mucins as discussed further below, but fewer such proteins were found in the fishes because we only identified one Muc2 in D. rerio and T. rubripes, two Muc5 in T. rubripes, and six Muc5 in D. rerio (SI Table 1).
Fig. 2.
Distribution of mucins. Shaded boxes indicate where a protein was found. Organisms are Homo sapiens (Hs), Mus musculus (Mm), G. gallus (Gg), X. tropicalis (Xt), T. rubripes (Tr), D. rerio (Dr), C. intestinalis (Ci), B. floridae (Bf), D. melanogaster (Dm), C. elegans (Ce), and N. vectensis (Nv). In the case where MV proteins have been indicated, a classification into Muc2, Muc5, etc. is not possible. A more detailed version of this table is found in SI Table 1.
The C. intestinalis proteins denoted MucMV3 (MV for “multiple VWDs”) and MucMV6 have D1-D4 domains that unambiguously assign them to the mucin/VWF/otogelin group of proteins (for MucMV3, which has a PTS domain, see Fig. 1 A and C). In vertebrate VWF and otogelins, the VWD domains occur together with VWA and AbfB domains, respectively. However, no proteins with these properties were identified in C. intestinalis. It therefore seems likely that the VWF and otogelin proteins are unique to vertebrates.
The Drosophila hemolectin protein (10, 11) and other insect orthologues have the VWD-VWE-TIL motif in common with mucins, and the Drosophila hemolectin protein also has F5/8 type C domains as well as a CysD domain as discussed below. We identified a protein related to hemolectin in B. floridae. The phylogenetic analysis of the D1, D2, D3, and D4 domains of hemolectin and the B. floridae proteins shows that both of these proteins belong to the mucin/otogelin/VWF group (Fig. 1C). Interestingly, the hemolectin protein has a domain structure very similar to a B. floridae protein, but the B. floridae protein has PTS domains inserted in a region that also contains multiple F5/8 type C domains (Fig. 1A). In B. floridae, therefore, the protein is highly similar in domain structure to a mucin, although mucins of higher animals do not have F5/8 type C domains. The B. floridae protein may thus be considered a hybrid between hemolectin and the gel-forming mucins MUC2 and MUC5AC/B in mammals.
The Origin of Gel-Forming Mucins Can Be Traced to Lower Metazoa.
VWD domains seem to be restricted to metazoa. A search among proteins predicted from available genome sequences of N. vectensis revealed two proteins with three consecutive VWD-VWE-TIL blocks, showing that this module was present already in this lower metazoan. In one of the proteins, inferred from analysis of genome sequence by using Genewise, three VWD-VWE-TIL blocks are followed by PTS and F5/8 domains, reminiscent of the B. floridae protein discussed above. In addition, the D1–D4 domains clearly map to the mucin group (Fig. 1C). Therefore, this protein has important characteristics of gel-forming mucins, and its presence in N. vectensis strongly suggests that gel-forming mucins were an early metazoan invention. The N. vectensis mucin-like protein has F5/8 domains, also found in mammalian SCO-spondin (12). However, the VWD domains of SCO-spondin do not seem to be phylogenetically related to the mucin-type VWD domains, and therefore, it seems unlikely that the N. vectensis protein is a SCO-spondin homologue.
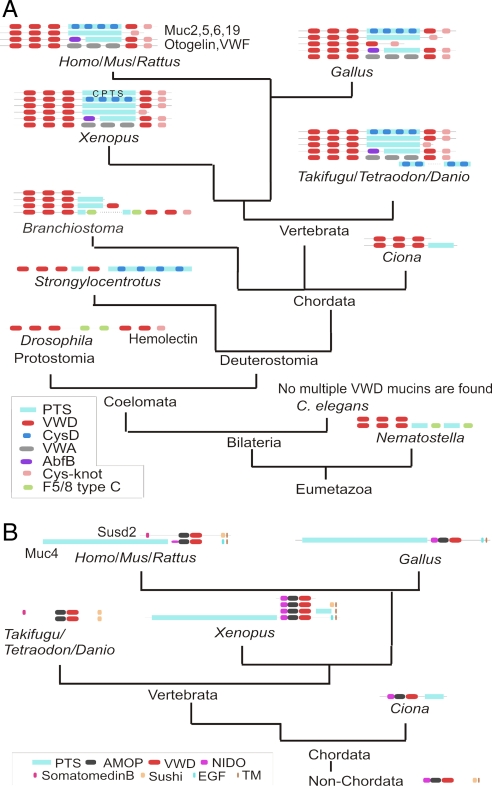
Our current view of the evolution of gel-forming mucins is summarized in Fig. 3A. The D1–D4 domains of the N. vectensis protein, insect hemolectin, S. purpuratus, B. floridae, and C. intestinalis mucin-like proteins, VWF, otogelin, and the gel-forming mucins are clearly related as judged by our analysis, and possibly the ancestor of these proteins was similar to the N. vectensis protein. Gel-forming mucins of the type observed in mammals have arisen by the combination of the VWD-VWE-TIL module with PTS, CysD, and cysteine-knot domains.
Fig. 3.
Phylogenetic distribution of gel-forming mucins (A) and Muc4 (B). The current phylogenetic distribution is shown in the context of metazoan taxonomy. Schematic domain structures are shown, and domains are not drawn to scale.
The Number of Gel-Forming Mucin Genes Has Increased Markedly in Xenopus.
The genes for the human and mouse gel-forming mucins MUC6, MUC2, MUC5AC, and MUC5B are in one locus, with this gene order, and the MUC6 gene has an orientation opposite to the others (SI Fig. 6). These mucins are evolutionarily related, as judged from sequence analysis and domain structure, and presumably arose by gene-duplication events (13). Proteins related to MUC2/5AC/5B/6 were identified in other vertebrates and were classified based on phylogenetic analysis (Fig. 1B and SI Fig. 5). In X. tropicalis and the fishes, however, 5ac and 5b cannot be distinguished. In C. intestinalis, Muc5 and Muc2 cannot be distinguished, and we have named these Ciona mucins MucMV1, MucMV2, etc. (SI Table 1).
Examination of the homologues in X. tropicalis now shows that many more proteins of the Muc2/6/5ac/5b type are found in this organism as compared with mammals. Thus, 16 Muc2 homologues and nine proteins of the Muc5 type were identified. (SI Fig. 7). Because we cannot classify the Muc5 proteins, we refer to them as Muc5a, Muc5b, Muc5c, etc. Twelve of the Muc5 and Muc2 proteins are localized to a contig covering 1.15 Mb of genomic sequence that seems to be related to the cluster in mammals, although the Xenopus cluster has a much more complex structure (SI Fig. 6).
Muc4 Evolved in Vertebrates by Combining NIDO-AMOP-VWD Domains with PTS Domains.
The Muc4 mucins form a group distinct from the other VWD-containing mucins, because they do not contain cysteines and have NIDO and AMOP domains N-terminal of VWD as shown in Fig. 1A. The only other protein with this arrangement is the sushi domain-containing protein Susd2 that lack PTS domains. A phylogenetic analysis of the AMOP and VWD domains of Susd2 homologues as well as Muc4 homologues (SI Fig. 8) confirms our classification of these proteins. The Susd2 protein has homologues in Xenopus and other vertebrates and in C. elegans, Drosophila, and Nematostella (Fig. 3B). On the other hand, a protein similar to human MUC4, with a PTS domain N-terminal of the NIDO-AMOP-VWD domains, seems first to appear in Xenopus (Fig. 3B). This finding, together with the close relationship of Susd2 and Muc4, gives support to the notion that MUC4 came about by the addition of a PTS domain to the NIDO-AMOP-VWD-TM arrangement from the sushi domain proteins, as previously suggested by Duraisamy et al. (14).
The Alternating PTS and CysD Domains Characteristic of MUC2/5 Type Mucins Are Present in S. purpuratus.
The CysD domain is a cysteine-rich domain (1, 13) that is adjacent to or within PTS regions as found in MUC2 and MUC5 from mammals. The CysD domain is also present in other proteins, such as the vertebrate cartilage intermediate layer protein (CILP) and the oikoplastic epithelium of tunicate (15). A profile HMM (Hidden Markov Model) using available CysD sequences (www.medkem.gu.se/mucinbiology/databases) was used to search for this domain in all available protein sequences. In mucin-like proteins from mammals as well as Xenopus and fish, it typically occurs C-terminal of the VWD-VWE-TIL block and adjacent to PTS domains. It is also found at this position in hemolectin where a PTS domain is lacking. In S. purpuratus, a protein has a PTS domain adjacent to CysD, and in C. intestinalis and B. floridae, there are multiple alternating PTS and CysD domains as found in the higher animals.
Nonvertebrate PTS Domains also Have a Repetitive Nature.
Mucins are characterized by PTS domains, and we have identified such domains in proteins from vertebrates as well as in lower animals. The PTS domains of mammalian mucins typically contain repeats as in human MUC1 and MUC2. This is also true for the vast majority of PTS domains identified in this work. Also the PTS domains in N. vectensis has a repetitive nature, most likely reflecting a common mechanism of recombination to produce a longer PTS domain. Detailed information on the repeats is available at www.medkem.gu.se/mucinbiology/databases. Although we have not found PTS domains as long as in the human mucins, some of the PTS mucins are exceptionally long, such as a D. rerio Muc5, with 2,571 aa and 61 repeats.
Gel-Forming Mucins in Xenopus Have Unusual PTS Domains Rich in Cysteines.
We noted that the majority of Muc2 proteins in X. tropicalis have unusual PTS domains. These domains (referred to as CPTS) contain cysteines, in contrast to PTS domains previously found in mucins. CPTS domains are also characterized by a repetitive structure, as exemplified in xMuc2g, with 92 repeat units of a sequence 46 aa in length (SI Figs. 7 and 9). We have not observed this type of Cys-rich PTS domains in any other organism. PTS domains of nine different X. tropicalis Muc2 proteins are also unusual because they are encoded by multiple exons, reminiscent of the gene structure for chicken Muc13 (8). Interestingly, none of the frog mucins with the Cys-rich PTS repeats (CPTS) have any CysD domains (SI Fig. 7). This suggests that CPTS and CysD domains present two different solutions to the problem of generating interactions between mucin chains.
SEA Domain Mucins Are Restricted to Vertebrates.
The human mucins MUC1, MUC3, MUC13, and MUC16 all contain SEA domains. SEA domains are also found in agrin, enterokinase (PRSS7), 63-kDa sea urchin sperm protein, HSPG2 (perlecan, heparan sulfate proteoglycan 2), the cell surface antigen 114/A10, HAT, a trypsin-like serine protease, IMPG1 and 2 (interphotoreceptor matrix proteoglycans 1 and 2), and in a number of proteins from Drosophila and C. elegans. However, by definition, only the mucins contain PTS domains.
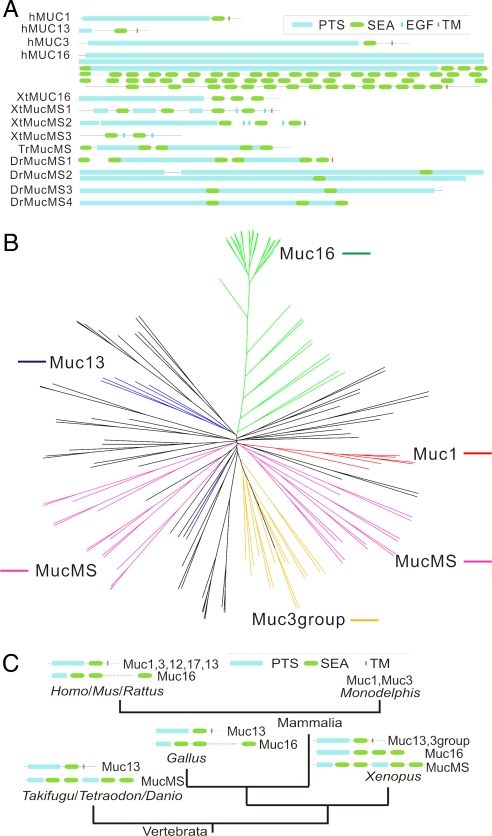
Searching genomes for SEA domains, we identified 149 SEA domains in 83 proteins. Among these proteins 39 were mucins with a total of 96 SEA domains. A marked expansion of SEA domains occurs in X. tropicalis and D. rerio that have a number of proteins with PTS and SEA domains. In addition, in X. tropicalis and the fishes, we identified a previously uncharacterized type of mucin-like protein that forms a rather large family. In this family, the PTS and SEA domains are alternating in a manner not seen in mammalian mucins, and we named these MucMS (MS for “multiple SEAs”). Selected proteins of this category are shown in Fig. 4A.
Fig. 4.
SEA domain proteins. (A) Domain structures of SEA domain-containing protein, including mucins and mucin-like proteins in Xenopus and fishes with alternating SEA and PTS domains. Proteins shown include previously known human mucins (hMUC) and human IMPG2, as well a previously uncharacterized type of mucins from T. rubripes (TrMuc), X. tropicalis (XtMuc), and D. rerio (DrMuc). (B) Phylogenetic tree of SEA domains. One hundred forty-nine VWD domains from 83 proteins were aligned by using ClustalW, and shown is the neighbor-joining tree derived by the same program. The mucin groups shown are Muc16 (green), Muc13 (blue), Muc1 (red), Muc3 (yellow), and the previously uncharacterized type of mucins with alternating PTS and SEA domains (pink). (C) Phylogenetic distribution of SEA domain mucins. The current phylogenetic distribution is shown in the context of metazoan taxonomy. Schematic domain structures are shown where domains are not drawn to scale. In the case of Monodelphis, only C-terminal parts of Muc1 and Muc3 have been identified, and the full domain structure is not known. Multiple SEA domains in the Homo and Gallus Muc16 have been left out.
Five different clusters of mucin-type proteins are identified from the phylogenetic analysis shown in Fig. 4B. One of them is Muc16, which is characterized by multiple SEA domains in human and mouse (3). A Muc16 homologue in chicken with multiple SEA domains has recently been described (8, 14). Here, we identified a homologue in X. tropicalis with three SEA domains. No Muc16 homologues could be identified in fishes or in any other more deeply branching animals (i.e., animals that are on branches closer to the root of the phylogenetic tree).
MUC1 homologues have previously been identified in primates, rodents, and cow (16), and here, we found a homologue also in opossum (M. domestica). However, there was no evidence of MUC1 in lower animals like chicken, frog, and fishes, suggesting that MUC1 appeared later in evolution than the other mucins.
The human MUC3, MUC12, and MUC17 mucin genes are found in a locus at the q22 region of chromosome 7. These mucins and their mouse homologues are described in SI Text and SI Fig. 10. We also identified a Muc3 homologue in opossum, but there was no evidence of additional paralogues (Muc12 and 17) as in human. In addition, a number of Muc3-like proteins are found in X. tropicalis, but no obvious Muc3 homologues could be identified in the fishes or in any other lower animal.
We recently reported Muc13 homologues in chicken and zebrafish (8) but could not find evidence of Muc13 in other chordates such as C. intestinalis. It would therefore seem that MUC13 is restricted to vertebrates.
The SEA domain is apparently an early metazoan invention because it occurs also in the deeply branching N. vectensis. SEA domain mucins of the type identified in mammals seem to have evolved in the vertebrate lineage because we could not observe obvious SEA domain mucin candidates in other phylogenetic groups (Fig. 4C).
The MUC1 SEA domain is autocatalytically cleaved, a reaction involving a serine hydroxyl (3). A GSX (where X can be V, I, or T) consensus sequence is believed to be critical for this catalytic activity. We noted that these sequence characteristics could be identified in all of the SEA domains of mucins except in the Muc16 group.
Discussion
A large number of mucin proteins were identified in a range of metazoan species and provided a comprehensive view of mucin phylogeny (Fig. 2). The PTS domains characteristic of all mucins are very poorly conserved in sequence and cannot be used to infer evolutionary history. Presumably, their sequence is less conserved because the exact location of serine and threonine residues acting as attachment sites for O-glycans is not crucial. A common theme is, however, the expansion of PTS domains through repeats, already observable in Nematostella. The length of the mucin PTS domains is probably very important for the properties of the mucus, but we do not know enough about selection mechanisms involved in the evolution of PTS domain. The PTS domains are typically encoded by one single exon, and their length exhibits allelic variation in humans. An interesting observation is that, in some species, each repeat is encoded by a separate exon. This will allow yet another level of regulation because the PTS length can be modified by alternative splicing.
We also discovered a previously uncharacterized type of PTS repeats containing cysteines in Xenopus. A primary function of gel-forming mucins is probably to generate protective net-like gels, and important for this function are cross-links between mucin chains. Cysteines of the CPTS domains are likely to play a role in this regard. We noted that CPTS and CysD domains are mutually exclusive in Xenopus mucins, suggesting that these two domains might have similar roles, i.e., to act as cross-linking sites. The presence of the CPTS domains as well as the larger number of mucins in Xenopus might reflect specific requirements in these animals. Frogs have mucus covering not only their internal organs, but also their skin. This might be related to the observation that frogs are more resistant to infections upon surgery, an observation that further illustrates the functional importance of mucins and mucus.
The gel-forming mucins are characterized not only by the PTS domains, but also by multiple VWD domains. In most cases, the VWD domain has adjacent VWE and TIL domains. An arrangement of three N-terminally located VWD-VWE-TIL modules is common and is known to be involved in oligomerization. The same module is found in proteins of the hemostasis or coagulation system of both insects (hemolectin) and vertebrates (VWF). These proteins lack a PTS domain but are indeed closely related to the gel-forming mucins. The common denominator for hemolectin, VWF, and the gel-forming mucins is their capacity to from polymers, a property that has to be well regulated in the cell to first take place at a later stage in the secretory pathway. Although the VWD domain was named after its presence in VWF, it is clear that this domain (as well as the VWD-VWE-TIL group of domains) was present in more ancestral proteins before the appearance of VWF. The important characteristics of a gel-forming mucins can be traced down to deeply branching organisms such as Nematostella. This suggests not only that the gel-forming mucins are ancestors to hemolectin and VWF but also that gel-forming mucins have a long evolutionary history.
Whereas the gel-forming mucins could be traced to early metazoan evolution, all of the membrane anchored mucins, both the mucins with SEA domains and the VWD-containing Muc4, seem to have appeared in the vertebrate lineage. It was previously known that the VWD domain is intimately linked to secreted mucin, but the discovery of a previously unrecognized family of secreted mucins (named MucMS) in Xenopus with repeated SEA domains interrupted by PTS domains emphasizes an important role also for the SEA domain in the context of mucins. The fact that the transmembrane mucins do not appear until the vertebrate lineage probably reflects a different functional role of these mucins as compared with the gel-forming mucins. This difference should be related to the role of the membrane-bound mucins in signaling and regulatory mechanisms that are specific to vertebrates, best illustrated by the MUC1 mucin (2) that first appeared among mammals.
The classification of mucins is not straightforward and has been subject of discussion (17). However, the mucin types discussed here are intimately related because they share localization to mucosal surfaces and have either VWD or SEA domains. The secreted gel-forming mucins with their VWD D1-D2-D3 PTS architecture are well defined. However, the “transmembrane” mucins are a more heterogeneous group of proteins, a group that also includes splice variants that are secreted. Instead, we prefer a classification based on their characteristic domains and suggest the names “SEA mucins” and “NIDO-AMOP-VWD mucins” (or “MUC4′”) for these categories of mucins that previously were listed under transmembrane mucins.
A comparison of proteins in mammals to homologues in more deeply branching animals shows that many mammalian proteins have evolved by a modification of domain architecture (18). This is also true for mucins. For example, in the MUC4 mucin the PTS domain is added to a NIDO-AMOP-VWD module present also in lower animals, and the later gel-forming mucins develop from simpler proteins with VWD-VWE-TIL and PTS domains, where the PTS domain is eventually expanded and CysD and cysteine knot domains are recruited. Interesting examples of protein domain evolution are also observed when comparing gel-forming mucins to mucin-like proteins without PTS domains such as hemolectin and VWF.
Materials and Methods
To identify proteins with VWD and SEA domains, such proteins were identified with hmmsearch or hmmpfam of the Hmmer package (http://hmmer.wustl.edu) and Pfam profiles of these domains obtained from Pfam (9, 19). All protein sequences available at the National Center for Biotechnology Information GenBank as well protein sequence data sets from ENSEMBL from organisms listed in Results were analyzed. All proteins thus identified were analyzed further with all profiles of Pfam as well as analyzed with respect to TM domains (20) and signal sequences (21). PTS domains were identified by using in house perl scripts using criteria previously described (7).
For phylogenetic analysis, VWD or SEA domain sequences were extracted and aligned by using ClustalW using default parameters. The bootstrapping option of ClustalW was used to generate bootstrapped trees. As an alternative, PHYLIP programs NEIGHBOR and PROTPARS were used to generate neighbor-joining and parsimony trees, respectively. One hundred replicates were analyzed for bootstrapping analysis, and CONSENSE was used to generate consensus trees.
A detailed description of genomic and protein sequence resources and computational methods are in SI Text, Materials and Methods.
Acknowledgments
T.L. was supported by the Swedish Knowledge Foundation through the Medical Bioinformatics PhD program at the Karolinska Institute (Stockholm, Sweden) and by a grant from the Sahlgren's Hospital (grant to Nils Lycke). This work was also supported by Swedish Research Council Grant 7461, the Swedish Cancer Foundation, and the Swedish Foundation for Strategic Research (MIVAC).
Abbreviations
- VWD
von Willebrand domain
- SEA
sea urchin sperm enterkinase agrin domain
- PTS
proline threonine serine-rich domain
- TIL
trypsin inhibitor-like.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705984104/DC1.
References
- 1.Perez-Vilar J, Hill RL. J Biol Chem. 1999;274:31751–31754. doi: 10.1074/jbc.274.45.31751. [DOI] [PubMed] [Google Scholar]
- 2.Hollingsworth MA, Swanson BJ. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 3.Macao B, Johansson DG, Hansson GC, Hard T. Nat Struct Mol Biol. 2006;13:71–76. doi: 10.1038/nsmb1035. [DOI] [PubMed] [Google Scholar]
- 4.Godl K, Johansson ME, Lidell ME, Morgelin M, Karlsson H, Olson FJ, Gum JR, Jr, Kim YS, Hansson GC. J Biol Chem. 2002;277:47248–47256. doi: 10.1074/jbc.M208483200. [DOI] [PubMed] [Google Scholar]
- 5.Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Buller HA, Dekker J, Van Seuningen I, Renes IB, et al. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Shi X, Chamankhah M, Visal-Shah S, Hemmingsen SM, Erlandson M, Braun L, Alting-Mees M, Khachatourians GG, O'Grady M, Hegedus DD. Insect Biochem Mol Biol. 2004;34:1101–1115. doi: 10.1016/j.ibmb.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 7.Lang T, Alexandersson M, Hansson GC, Samuelsson T. Glycobiology. 2004;14:521–527. doi: 10.1093/glycob/cwh066. [DOI] [PubMed] [Google Scholar]
- 8.Lang T, Hansson GC, Samuelsson T. BMC Genomics. 2006;7:197. doi: 10.1186/1471-2164-7-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al. Nucleic Acids Res. 2004;32:D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goto A, Kumagai T, Kumagai C, Hirose J, Narita H, Mori H, Kadowaki T, Beck K, Kitagawa Y. Biochem J. 2001;359:99–108. doi: 10.1042/0264-6021:3590099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesch C, Goto A, Lindgren M, Bidla G, Dushay MS, Theopold U. Dev Comp Immunol. 2007 doi: 10.1016/j.dci.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Meiniel O, Meiniel A. Brain Res Rev. 2007;53:321–327. doi: 10.1016/j.brainresrev.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Desseyn JL, Aubert JP, Porchet N, Laine A. Mol Biol Evol. 2000;17:1175–1184. doi: 10.1093/oxfordjournals.molbev.a026400. [DOI] [PubMed] [Google Scholar]
- 14.Duraisamy S, Ramasamy S, Kharbanda S, Kufe D. Gene. 2006;373:28–34. doi: 10.1016/j.gene.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 15.Spada F, Steen H, Troedsson C, Kallesoe T, Spriet E, Mann M, Thompson EM. J Biol Chem. 2001;276:20624–20632. doi: 10.1074/jbc.M100438200. [DOI] [PubMed] [Google Scholar]
- 16.Spicer AP, Duhig T, Chilton BS, Gendler SJ. Mamm Genome. 1995;6:885–888. doi: 10.1007/BF00292441. [DOI] [PubMed] [Google Scholar]
- 17.Dekker J, Rossen JW, Buller HA, Einerhand AW. Trends Biochem Sci. 2002;27:126–131. doi: 10.1016/s0968-0004(01)02052-7. [DOI] [PubMed] [Google Scholar]
- 18.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 19.Eddy SR. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 20.Sonnhammer EL, von Heijne G, Krogh A. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 21.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]