Abstract
The male accessory glands (MAGs) of many insect species produce and secrete a number of reproductive proteins collectively named Acps. These proteins, many of which are rapidly evolving, are essential for male fertility and represent formidable modulators of female postmating behavior. Upon copulation, the transfer of Acps has been shown in Drosophila and other insects to trigger profound physiological and behavioral changes in females, including enhanced ovulation/oviposition and reduced mating receptivity. In Anopheles gambiae mosquitoes, the principal vectors of human malaria, experimental evidence clearly demonstrates a key role of MAG products in inducing female responses. However, no Acp has been experimentally identified to date in this or in any other mosquito species. In this study we report on the identification of 46 MAG genes from An. gambiae, 25 of which are male reproductive tract-specific. This was achieved through a combination of bioinformatics searches and manual annotation confirmed by transcriptional profiling. Among these genes are the homologues of 40% of the Drosophila Acps analyzed, including Acp70A, or sex peptide, which in the fruit fly is the principal modulator of female postmating behavior. Although many Anopheles Acps belong to the same functional classes reported for Drosophila, suggesting a conserved role for these proteins in mosquitoes, some represent novel lineage-specific Acps that may have evolved to perform functions relevant to Anopheles reproductive behavior. Our findings imply that the molecular basis of Anopheles female postmating responses can now be studied, opening novel avenues for the field control of these important vectors of human disease.
Keywords: reproduction, malaria, mating, Acp, seminal fluid
Anopheles gambiae mosquitoes, the major vectors of human malaria, transmit Plasmodium parasites with a remarkable efficiency. The ability of this mosquito species to function as a malaria vector depends on multiple factors, including a strong preference for humans as hosts for blood feeding, a high susceptibility to parasite infections, a long lifespan, and a high reproductive rate. The identification of Anopheles genes regulating vectorial capacity is therefore a research priority, because their manipulation would provide important clues for the development of novel vector control measures aimed at fighting this devastating disease.
An. gambiae females, similar to females from a number of other insect species, undergo a series of changes upon insemination that dramatically modify their physiology and behavior: the phase of the flight activity rhythm shifts from crepuscular to nocturnal, ovulation and oviposition are triggered, and lifetime refractoriness to further copulation is induced, with multiple matings occurring in only a small percentage of individuals (1). In the fruit fly Drosophila melanogaster postmating responses are modulated by components of the seminal fluid, produced primarily by the male accessory glands (MAGs) and transferred to females during mating. Besides stimulating egg laying and triggering reduced sexual receptivity, MAG secretions have been shown to induce expression of immune peptides and reduction of female lifespan (reviewed in refs. 2–4). MAG products therefore are essential modulators of female behavior and represent important targets for biological and genetic control of insect pests (4).
The major components of MAG secretions are proteins, collectively named Acps. Thus far 59 of the 112 Acps postulated to be present in the D. melanogaster genome have been identified, and a small number has been shown to play a role in reproduction (5–11). The majority of these genes encode proteins with secretory signal peptides and exhibit male-specific expression that is highly enriched in MAGs, two criteria used to define bona fide Acps (6, 11). They include some of the most rapidly evolving genes in the Drosophila genome (10), and accordingly numerous attempts to identify their orthologues in other insects have failed (12–14).
In An. gambiae mosquitoes a crucial role for MAG secretions in modulating female postmating responses is well established. Experiments based on normal and forced copulation demonstrated that substances from the MAGs are necessary and sufficient to induce refractoriness to subsequent inseminations in females and to stimulate oviposition (15, 16). Females mated to males with degenerate testes and accessory glands failed to oviposit and readily remated, whereas females mated to males with degenerate testes but fully developed accessory glands laid (unfertilized) eggs and did not remate (15, 16). Unlike other insect species in which Acps are introduced into females as fluid secretions, MAG products in Anopheles are transferred during mating mainly as a solid mass (called the mating plug) (17) that slowly dissolves in the female reproductive tract. The means of delivery of MAG peptides in An. gambiae appears to be essential for their function because crude MAG homogenates do not produce measurable effects on mating refractoriness when injected directly into females (18). Despite the unquestionable importance of Anopheles mosquitoes for human health and the potential for their control presented by the manipulation of female postmating responses, no Anopheles MAG gene has been identified.
Here we report on the identification of 46 genes from the MAGs of An. gambiae mosquitoes, including the putative orthologues of 25 D. melanogaster Acps. The majority of these genes are specifically expressed in the male reproductive tract and show a low level of homology with their Drosophila orthologues. Comparative structural analysis suggests that MAG proteins share similar functions in the two insect species. However, a number of Anopheles-specific Acps are found, mostly organized in a densely populated cluster on chromosome arm 3R. These findings increase our knowledge of the molecular components of reproductive behavior in mosquitoes and expand the field of research on the role of MAG proteins, so far principally focused on studies in the Drosophila genus, to other medically and economically important insect species.
Results
Identification of Acp Genes in An. gambiae.
With the aim to identify male genes modulating female postmating responses, the An. gambiae genome was searched for orthologues of 59 known D. melanogaster Acps (5–10). Initial standard sequence similarity (BLAST) and sequence colinearity (synteny) searches identified in Anopheles 20 putative Acps orthologues, mainly encoding large proteins. RT-PCR showed that 15 of these genes, similar to their Drosophila orthologues (www.flyatlas.org), were expressed in most tissues analyzed, which included the MAGs, testes, male carcasses (i.e., males deprived of their internal reproductive organs), and whole females (Table 1). The remaining five genes identified by the BLAST analysis were not detected in MAGs [supporting information (SI) Table 2].
Table 1.
An. gambiae genes expressed in the MAGs
| Anopheles gene | Drosophila homologue | Identity, % | Length, aa | Functional class | Method | Chr |
|---|---|---|---|---|---|---|
| MAG-only | ||||||
| 06418 | CG17575 | 27 | 266 | CAP | M | 2L |
| 06585 | Acp63F | 29 | 83 | Protease inhibitor | M | 2L |
| 06587 | Acp62F | 27 | 91 | Protease inhibitor | M | 2L |
| (12682) | CG15353* | 30 | 48 | C | 3R | |
| 09353 (12680) | Msopa | 21 | 81 | M | 3R | |
| 09358-RB | Nplp4-PA* | 24 | 80 | C | 3R | |
| 09358-RA (12830) | Nplp4-PA* | 25 | 80 | C | 3R | |
| 09359 | Mst57Da | 25 | 71 | M | 3R | |
| 12718† | CG15065* | 26 | 52 | C | 3R | |
| 09362 | CG6409* | 19 | 225 | C | 3R | |
| 09365† | CG5793* | 42 | 309 | Transport protein | C | 3R |
| 09367 | Acp26Ab | 22 | 91 | M | 3R | |
| 09368 | CG14770* | 21 | 129 | C | 3R | |
| 09369 | Acp53Ea | 19 | 100 | M | 3R | |
| 09370 (12706) | Acp53Ea | 22 | 98 | M | 3R | |
| 09371 | CG14302* | 25 | 72 | C | 3R | |
| 09373 | CG16707‡ | 22 | 210 | C | 3R | |
| MAG, T | ||||||
| 09352 (12681) | Acp70A | 27 | 56 | M | 3R | |
| 09354 | Mst57Da | 25 | 71 | M | 3R | |
| 09355 | Dro-PA* | 26 | 55 | C | 3R | |
| 09356 | Mst57Da | 24 | 71 | M | 3R | |
| 09357 | Dro-PA* | 26 | 55 | C | 3R | |
| 09360 (12807) | CG13230* | 17 | 68 | C | 3R | |
| 09361 | Acp95EF | 25 | 54 | M | 3R | |
| 09372 | CG32726* | 21 | 69 | C | 3R | |
| MAG, T, F | ||||||
| COEBE4D | EST-6 | 31 | 557 | Carboxylesterase | B | 2L |
| COEBE1D | EST-6 | 32 | 570 | Carboxylesterase | B | 2L |
| 06583 | Acp63F | 22 | 81 | Protease inhibitor | M | 2L |
| 01424† | CG5520 | 62 | 959 | Chaperone | B | 2R |
| 04428† | CG3359 | 39 | 822 | Cell adhesion | B | 2R |
| 09429 | Anp | 26 | 75 | M | 3R | |
| MAG, RB, F | ||||||
| 06581 | Acp62F | 23 | 94 | Protease inhibitor | M | 2L |
| 06586 | Acp62F | 24 | 94 | Protease inhibitor | M | 2L |
| 03083 | CG17097 | 39 | 428 | Lipase | B | 2R |
| 09364 | CG5793* | 52 | 310 | Transport protein | C | 3R |
| Ubiquitous | ||||||
| 05246 | CG9334 | 38 | 395 | Protease inhibitor | B | 2L |
| 07049 | CG10433 | 37 | 136 | β-Defensin | B | 2L |
| 07088 | CG2852 | 72 | 202 | Isomerase | B | 2L |
| 07491 | CG4670 | 27 | 661 | Redox | B | 2L |
| SRPN9 | CG10956 | 18 | 416 | Protease inhibitor | B | 2R |
| Calreticulin | Crc | 71 | 406 | Chaperone | B | 2R |
| TEP15 | CG10363 | 34 | 1422 | α2-Macroglobulin | B | 3R |
| 08822† | CG9847 | 62 | 175 | Isomerase | B | 3R |
| 08968 | CG31704 | 42 | 63 | Protease inhibitor | B | 3R |
| 09363† | CG11466* | 36 | 508 | Cytochrome P450 | C | 3R |
| 09842 | CG8194 | 35 | 312 | Ribonuclease | B | 3R |
The table contains 46 An. gambiae genes that are expressed exclusively or abundantly in the MAGs and their putative D. melanogaster homologues. Genes are grouped according to their expression profile, as determined by RT-PCR. T, testes; RB, rest of the body (whole males deprived of reproductive apparatus); F, whole females. The percentage of identity between putative homologues and the length of the Anopheles proteins are provided, as are their putative function when determined (see SI Table 3). The chromosomal arm on which genes are localized is indicated. The methods used for the initial identification of the Anopheles homologues are indicated (M, MatLab; B, BLAST searches; C, cluster on chromosome arm 3R). The Anopheles genes in parentheses represent putative alleles from an alternative haplotype. Gene 12682 is in parentheses because in Ensembl it is located on an unmapped scaffold, whereas our analysis predicts its localization on chromosome arm 3R. Anopheles gene 12718 is described with the first five digits of the old AgamP3 Ensembl identifier (ENSANGG000000012718) because it has not been assigned a new identifier in the AgamP3.4 gene build. 09358-RA and 09358-RB are labeled in AgamP3.4 as alternative transcripts of the same gene; however, they encode two separate genes and therefore are identified here by their transcript numbers. Chr, chromosome arm.
*Drosophila genes that are not known to be expressed in the male accessory glands in the fruit fly.
†Anopheles genes whose full-length sequence has been reconstructed (using Fgenesh, Fgenesh+, Wise2, and Genescan gene-finding software).
‡ This result was not confirmed by reverse BLAST analysis.
This initial analysis did not allow the identification of the orthologues of any of the male-specific Acps that in Drosophila trigger female postmating behavioral changes. To significantly improve the depth of our bioinformatics analysis, homology searches were performed by using the Smith–Waterman and Needleman–Wunsch approaches, as implemented in the Bioinformatics toolbox in the Matlab programming environment (for a detailed explanation of the bioinformatics analysis procedure see Materials and Methods and SI Appendices 1 and 2). Remarkably, this analysis identified 16 Anopheles genes abundantly expressed in MAGs, 12 of which are specifically expressed in the male reproductive tract, encoding the homologues of 11 additional Drosophila Acps (Table 1). These genes, encoding proteins <100 aa (with one exception), show very low identity (19–29%) to their Drosophila homologues, demonstrating the power of this bioinformatics analysis (Table 1). An additional seven putative orthologues instead could not be detected in MAGs but in some cases were detected in the testes or other male tissues (SI Table 2).
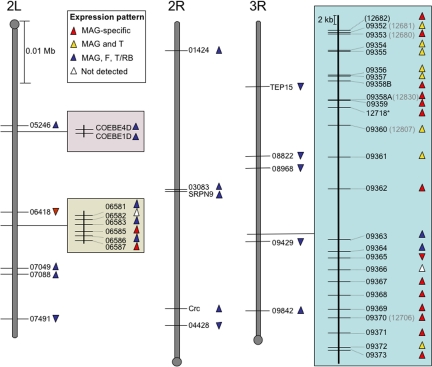
Several of the An. gambiae Acps identified were organized in a cluster on chromosome arm 3R (Fig. 1, blue-shaded area). A manual annotation of the region encompassing this cluster uncovered the presence of 15 additional genes, all encoding proteins with predicted signal peptides. RT-PCR analysis revealed all 15 genes to be highly expressed in MAGs, nine being MAG-specific and four being restricted to the male reproductive tract (MAGs and testes) (Table 1 and Fig. 1). The Drosophila orthologues of these genes were identified by using the Matlab programming environment as above and did not encode additional known Acps.
Fig. 1.
Chromosomal organization of 46 genes expressed in the MAGs of An. gambiae. These genes are localized on chromosome arms 2L, 2R, and 3R, which are indicated by gray bars with the centromere marked by a circle. Chromosome arms are drawn to scale. Direction of the triangles reflects orientation on the chromosomal strands, and the color is indicative of pattern of expression, as specified in the key. T, testes; RB, male carcasses without male reproductive organs; F, whole females. The shaded boxes next to chromosome arms 2L and 3R highlight the presence of gene clusters. The five-digit numbers refer to the vector base identifier (AgamP3.4 gene build), omitting the AGAP code and the first digit (i.e., 06418 refers to AGAP006418). (See Table 1 legend.)
In total, these analyses identified 46 genes showing strong expression in the MAGs of Anopheles, 25 being male reproductive tract-specific (17 specifically expressed in MAGs and eight restricted to MAGs and testes) (Table 1 and Fig. 1). Among these genes are the homologues of 25 Drosophila Acps, corresponding to >40% of the fruit fly genes analyzed here.
Male Reproductive Tract-Specific Acps.
The 25 genes specifically expressed in the male reproductive tract identified by bioinformatics analysis and manual annotation meet the stringent criteria that define bona fide Acps in Drosophila (6). All 25 genes encode proteins with predicted signal peptides, exhibit expression profiles restricted to the male reproductive tract and highly enriched in the MAGs, and lack any previously assigned non-Acp function. Importantly, 14 of these An. gambiae Acps are homologues of Drosophila Acps, showing low levels of identity (19–29%), and none could be identified by standard BLAST or synteny analysis (Table 1 and SI Appendices 1 and 2).
Included among these 25 genes are the homologues of Drosophila Acps implicated in the induction of female postmating responses. We have identified a putative orthologue of Acp70A, commonly referred to as sex peptide, which is by far the best-characterized Acp in Drosophila. Sex peptide has been shown to induce mating refractoriness, trigger ovulation, elicit feeding behavior, cause induction of the immune response, and reduce female lifespan (2, 4, 19). Like its identified An. gambiae orthologue, 09352 (SI Appendix 1), sex peptide is primarily expressed in MAGs but is also found in the testes at low levels (6). [Note that the An. gambiae genes are referred to with the last five digits of the VectorBase identifiers (i.e., 06418 refers to AGAP006418).]
An additional An. gambiae male-specific Acp is a homologue of the Drosophila Acp62F, a cysteine-rich trypsin-like protease inhibitor that has been demonstrated to be toxic to females when ectopically expressed (20). Allelic variation in Acp62F in males is associated with their success in sperm competition and the fecundity of their mating partners (21). The Anopheles homologue of Acp62F (06587) is clustered with the homologue of another Drosophila Acp, Acp63F, on chromosomal arm 2L (06585). This cluster contains three additional closely related genes expressed in MAGs and also in females (Table 1 and Fig. 1, green-shaded area).
Allelic variation in two other Drosophila Acps, Acp26Ab and Acp53Ea, for which putative orthologues were identified in our screen, is associated with sperm defense, the ability of an ejaculate present in a female to resist displacement by a second ejaculate (22). Among the Drosophila Acps for which we found male-specific orthologues in Anopheles, Acp70A, Acp62F, Acp26Ab, Msopa, and CG17575 are transferred to the female during copulation and enter the female circulatory system shortly after transfer (23).
A total of 12 male-specific Acps show similarity to Drosophila proteins with either unknown function or not known as MAG proteins. Two highly similar genes (09358-RB and 09358-RA) are related to the neuropeptide hormone Neuropeptide-like precursor 4 (Nplp4) (Table 1). A further two genes, 09355 and 09357, which are 100% identical to each other, are related to the antimicrobial peptide Drosocin (Dro-PA) (Table 1). In Drosophila, Drosocin is a proline-rich peptide, highly induced in the fat body by bacterial challenge, which upon mating is also found in the oviduct of egg-laying females (24). Another Anopheles gene, 09368, shows similarity to CG14770, which has not been described as an Acp in Drosophila but is up-regulated in MAGs (www.flyatlas.org) and has an orthologue in the medfly Ceratitis capitata, which has also been shown to be MAG-specific (12) (Table 1).
The striking majority (22 of 25) of the male-specific genes are organized in the 3R cluster. This region, spanning 48 kb, contains exclusively MAG-expressed genes (Fig. 1, blue-shaded area) and appears to have evolved as a male “fertilization island” in the An. gambiae genome. The presence of this island represents a major difference with the organization of Acps in Drosophila, where, although some clustering exists in the form of tandem duplications, genes are predominantly dispersed throughout the genome. Several groups of paralogues are found in the cluster, indicating that some of these genes are likely to be the result of recent gene duplication. Large clusters of coordinately expressed genes have been documented in Drosophila in the case of genes coexpressed in spermatogenesis and coregulated during circadian cycles (25). The presence of this highly populated cluster suggests that coregulation of these MAG genes may be important for male reproductive biology.
Comparative Structural Modeling of An. gambiae Acps.
Because sequence similarity does not necessarily imply functional conservation, we used comparative structural analysis to help elucidate the function of 22 Anopheles Acps (SI Table 3). As expected, most of these genes fall in the same protein classes as their Drosophila orthologues, as described below.
CAP (CRISP/Antigen5/PR-1) Superfamily.
One Anopheles male-specific Acp, 06418, contains 10 conserved cysteine residues in the N-terminal CAP and hinge domains characteristic of members of the cysteine-rich secretory protein family (CRISPs), but like its Drosophila orthologue CG17575 lacks the ICR/CRD domain of true CRISPs. The 3D structure reveals two conserved histidine residues (H119 and H184) located in a pocket that likely form the Ca2+-binding site characteristic of CRISPs (SI Fig. 3A). CAP proteins lacking the ICR/CRD domain are known to function as sperm chemoattractants and inhibitors of sperm–egg fusion (26).
Lipid Transport Proteins (CRAL-TRIO Domain Proteins).
Two proteins identified here belong to the CRAL-TRIO family: the male-specific Acp 09365, and 09364, which is also expressed in females. CRAL-TRIO domain-containing proteins are characterized by large hydrophobic pockets thought to be involved in the transport of lipids or other small hydrophobic molecules. Both mosquito proteins thread onto human α-tocopherol transfer protein and retain large hydrophobic domains (SI Fig. 3B and SI Table 3). Lipids transferred to females in the ejaculate are known to modulate the postmating response in Drosophila (27).
Protease Inhibitors.
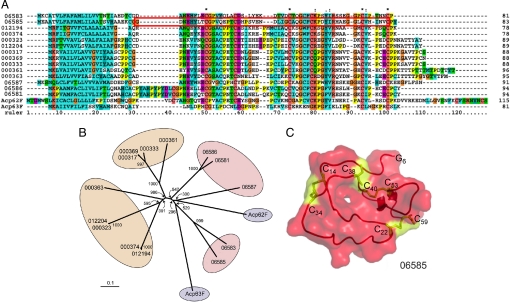
A total of eight genes thread to serine protease inhibitors (serpins), compared with the seven serpins found among Drosophila Acps (28). Serpins have been shown to have a role in male fertility in many mammalian species (29). The eight genes found here include five protease inhibitors, homologous to Acp62F and Acp63F (Fig. 1, green-shaded area, and Fig. 2), that belong to a small class of serpins with unique structural features (20). Fig. 2C shows a 3D model for 06585, a homologue of Acp63F, with the highly conserved cysteine residues forming disulfide bridges highlighted. Acp63F was not assigned to a functional class in previous studies in Drosophila (28). Interestingly, serpins related to Acp62F and Acp63F have undergone a considerable gene expansion in another mosquito species, the dengue and yellow fever vector Aedes aegypti. Based on the first draft of the recently assembled genome, a cluster can be identified containing nine highly related Aedes serpins showing a good degree of conservation with the Anopheles and Drosophila homologues (Fig. 2 A and B). Three additional protease inhibitors, the serpins 05246 (SRPN10) (30) and SRPN9 (http://cegg.unige.ch/insecta/immunodb), and a Kazal-type serpin (08968), are also expressed in the Anopheles MAGs (SI Table 3).
Fig. 2.
Phylogenetic analysis of serine protease inhibitors in three insect species. (A) Multiple protein sequence alignment for the D. melanogaster (Acp62F and Acp63F), An. gambiae (06587, 06586, 06585, 06583, and 06581), and Ae. aegypti (identified by the last six digits of their Ensembl entry codes omitting the AAEL code: 012194, 000374, 000323, 012204, 000317, 000369, 000333, 000361, and 000363) homologues generated by using Clustal X and Clustal W. Conserved cysteine residues are indicated by an asterisk. The red box in the sequence of 06585 specifies the region used for the 3D model shown in C. (B) Unrooted phylogenetic tree was constructed by the neighbor-joining method based on the sequence alignment as above. Anopheles genes are shaded in pink, Drosophila genes are shaded in blue, and Aedes genes are shaded in orange. The bootstrap values of 1,000 replicates are indicated. The scale bar represents the amino acid divergence. (C) Three-dimensional model of the Anopheles putative serine protease inhibitor 06585. The cysteine residues engaged in disulfide bridges and the free Cys-5g are shown in ball-and-stick form in yellow.
α/β-Hydrolases.
Two classes of hydrolase were identified in the MAGs of An. gambiae: one lipase and two carboxylesterases. Lipases are postulated to provide energy to sperm by hydrolysis of triglycerides (8). Like most lipases expressed in the MAGs of D. melanogaster, the Anopheles 03083 threads onto an acid lipase (SI Table 3), and it contains the active site (consisting of a catalytic triad and an oxyanion hole) characteristic of lipases (SI Fig. 3 E and F). Two carboxylesterase genes (COEBE4D and COEBE1D), clustered on chromosome 2L, are homologues of Drosophila EST-6 (SI Fig. 3D). EST-6 in Drosophila is expressed in the male genitalia and transferred to the female in the semen during mating, influencing egg-laying behavior and possibly receptivity to remating (31).
Immunity-Related Proteins.
Three antimicrobial peptides are found among the An. gambiae Acps identified here, although we could not generate 3D models for these proteins. It has been suggested that antimicrobial peptides found in male genitalia protect the male ejaculate or the female reproductive tract from microbial infections (32, 33). In addition to the Drosocin-like peptide described above, we found a gene encoding a protein with a β-defensin domain (07049) and the orthologue of Andropin (09429) (Table 1 and SI Appendix 1). Defensins are small cationic peptides involved in the immune defense against Gram-positive bacteria that are normally present in the female reproductive tract, whereas Andropin in Drosophila is active against both Gram-positive and Gram-negative bacteria and is highly enriched in MAGs, with lower expression levels also in testis.
Chaperones.
We found four MAG-expressed genes encoding proteins predicted to be involved in protein folding: two peptidylprolyl isomerases and two chaperone proteins (SI Table 3). Peptidylprolyl isomerases catalyze the conversion of peptide bonds with the amino acid proline from trans to cis thereby facilitating protein folding. One of these isomerases, 07088, is predicted to be a secreted cyclophilin-like protein and retains the cyclosporine-binding site of human cyclophilin B (SI Fig. 3C). The other three proteins from this group [08822 (an FKBP-like isomerase), 01424 (an hsp90-like protein), and a previously characterized calreticulin] all have the predicted C-terminal endoplasmic reticulum targeting sequence HDEL. However, such proteins can escape endoplasmic reticulum retention into the secretory pathway and have been implicated to have a role in reproduction by acting as intercellular signal molecules facilitating sperm–egg interaction (34, 35).
Redox Proteins.
Two redox proteins are expressed in MAGs. One, 09363, is a previously characterized microsomal cytochrome P450 (CYP9M1), and the other, 07491, is a thioredoxin-like protein belonging to an antioxidant class of proteins involved in protection from oxidative stress.
Discussion
We have achieved the identification of the first MAG genes from a mosquito vector of human malaria. The 46 An. gambiae Acps found here, 25 of which are male reproductive tract-specific, encode the homologues of 25 Drosophila known Acps, bridging the gap in knowledge between Anopheles and Drosophila. This result is remarkable considering that many Acps are rapidly evolving and diverge extensively among related insect species. Indeed, with the exception of the large proteins, most Anopheles Acps identified here show very low homology to their Drosophila orthologues, ranging from 19% to 29%.
We estimate that the genes identified here correspond to the majority of male reproductive tract-specific Acps present in Anopheles. The An. gambiae Affymetrix microarray database, available at www.angaged.bio.uci.edu, contains a total of 32 nonredundant Anopheles genes that potentially meet the stringent criteria for genuine Acps described above (6, 11), because they show a predicted signal peptide, their transcripts are enriched at least 5-fold in males than in female at any stage, and they do not have an assigned non-Acp function. Twenty of these 32 genes (therefore 62.5%) correspond to the male-specific Acps identified in our bioinformatics and molecular analyses (Table 1 and Fig. 1). The remaining five male-specific Acps do not have corresponding probe sets. Considering that the additional male-specific genes found in the above database will not necessarily be all expressed in the MAGs, we can conservatively estimate that we have identified two-thirds of bona fide Acps in An. gambiae. Future functional analyses are needed to clarify the role of single Acps in modulating female postmating behavior. However, given the number of Anopheles Acps identified here, it is reasonable to believe that indeed at least some of these proteins are transferred to females and perform reproductive functions.
In our analysis we found proteins belonging to a series of functional classes described for Drosophila Acps, including lipases, chaperones, serpins, and antimicrobial peptides (Table 1 and SI Table 3). However, some differences were found, and a number of Anopheles MAG genes have homologues in Drosophila that are not known as Acps. The microarray database of Drosophila available at www.flyatlas.org shows that transcripts of these fruit fly genes are not enriched in MAGs, suggesting that the An. gambiae homologues may represent lineage-specific Acps that have evolved to perform functions relevant to mosquito fertility. It is tempting to speculate that such proteins could account for the differences in postmating behavior observed between the two insect species, for instance the lifelong refractoriness of most An. gambiae females compared with the resumption of mating behavior observed in D. melanogaster 7 days after mating. The collection of Anopheles Acp genes identified here constitutes therefore an important reference set of mosquito male fertility genes that will undoubtedly benefit future analyses in other insect species and may provide novel leads for vector control strategies based on the manipulation of important female postmating responses such as oviposition and remating behavior.
Materials and Methods
Identification of the Anopheles Homologues of Drosophila Acps.
Selected D. melanogaster Acps were initially subjected to BLAST searches at NCBI (www.ncbi.nlm.nih.gov/BLAST) and at Ensembl (www.ensembl.org), at both the protein level and the DNA level. For all annotated Drosophila Acp genes we also analyzed the genome colinearity (synteny) data between Drosophila and Anopheles (36) present in AnoEST (http://web.bioinformatics.ic.ac.uk/AnoEST/anoest.php), and the automatic clustering of orthologues and in-paralogues from Inparanoid (http://inparanoid.sbc.su.se). For more stringent and sensitive bioinformatics analyses, homology searches were performed by using the Smith–Waterman and Needleman–Wunsch approaches, as implemented in the Bioinformatics toolbox in the Matlab programming environment (Matlab version 7.0.4), to identify homology between a selected set of Acp proteins from different Drosophila species (D. melanogaster, Drosophila mauritiana, Drosophila simulans, Drosophila sechellia, Drosophila subobscura, Drosophila madeirensis, and Drosophila pseudoobscura, downloaded from Swissprot, http://expasy.org/sprot) and the An. gambiae superset of translated sequences from known or novel gene predictions, as downloaded from Ensembl (database release version 42.3e, February 2006). Alignment tests were done by using different scoring matrices: generally the best results were obtained by using BLOSUM as scoring matrix, with a percent identity level of 62. Various penalties were assigned for opening or extending a gap during sequence alignment. Each run of the application resulted in a list of candidate amino acid sequences in the Anopheles genome that were subjected to reciprocal best-hit analysis against the D. melanogaster genome using the same parameters used in the initial similarity searches. For examples and references of the bioinformatics searches, see SI Appendices 1 and 2. The Matlab script that was used to obtain the results and the input data are readily available upon request.
Mosquito Dissections and RT-PCR Analysis.
Tissues from MAGs, testes, and male carcasses lacking the genitalia were dissected in 1× PBS solution, and total RNA was extracted from these tissues as well as from whole adult An. gambiae female mosquitoes (G3 strain) using TRI Reagent (Helena Biosciences, Gateshead, U.K.), according to the manufacturer's instructions. The RNA was reverse transcribed by using oligo d(T)s and M-MLV Reverse Transcriptase (Invitrogen, Paisley, U.K.) following standard protocols. RT-PCRs were carried out by using the HotStarTaq DNA Polymerase (Qiagen, Valencia, CA) as described by the manufacturer, using primer sets specific for each gene. A total of 35 cycles per reaction were run. At least two replicates were performed for each gene analyzed in each tissue. As a control for the specificity of our amplifications, we used primers for the testis-specific β2-tubulin gene (37) on MAGs and testes cDNA samples. The ribosomal gene S7 was used as a positive control for all cDNA samples. In five cases, the amplified products were cloned and sequenced to confirm their identity. In all cases the sequences obtained corresponded to the original Ensembl gene prediction.
Phylogenetic Analysis of Serpins.
The selected Drosophila, Anopheles, and Aedes amino acid sequences were subjected to multiple alignments by means of the Clustal W (www.ebi.ac.uk/Tools/clustalw) and Clustal X (Version 1.83) algorithms. Phylogenetic analysis was performed by using TreeView software (Version 1.6.6) (38). A phylogenetic tree was constructed by the neighbor joining method by using p-distance estimates and tested by using the interior-branch test. Reliability of each node was assessed with 1,000 bootstrap replications.
Generation of 3D Models.
Three-dimensional templates were generated for 22 of the 46 Anopheles Acp proteins. See the legend of SI Table 3 for details.
Supplementary Material
Acknowledgments
We thank Rebecca Armson for initial efforts on this project, Nicola Senin for help with the Matlab programming environment, and Elena Levashina for invaluable suggestions. This work was supported by a Wellcome Trust VIP Award and a Medical Research Council Career Development Award, Agreement ID 78415, file no. G0600062 (to F.C.).
Abbreviation
- MAG
male accessory gland.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703904104/DC1.
References
- 1.Tripet F, Toure YT, Dolo G, Lanzaro GC. Am J Trop Med Hyg. 2003;68:1–5. [PubMed] [Google Scholar]
- 2.Kubli E. Cell Mol Life Sci. 2003;60:1689–1704. doi: 10.1007/s00018-003-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfner MF. Insect Biochem Mol Biol. 1997;27:179–192. doi: 10.1016/s0965-1748(96)00084-7. [DOI] [PubMed] [Google Scholar]
- 4.Chapman T, Davies SJ. Peptides. 2004;25:1477–1490. doi: 10.1016/j.peptides.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 5.Chen PS, Stumm-Zollinger E, Aigaki T, Balmer J, Bienz M, Bohlen P. Cell. 1988;54:291–298. doi: 10.1016/0092-8674(88)90192-4. [DOI] [PubMed] [Google Scholar]
- 6.Mueller JL, Ravi Ram K, McGraw LA, Bloch Qazi MC, Siggia ED, Clark AG, Aquadro CF, Wolfner MF. Genetics. 2005;171:131–143. doi: 10.1534/genetics.105.043844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simmerl E, Schafer M, Schafer U. Insect Biochem Mol Biol. 1995;25:127–137. doi: 10.1016/0965-1748(94)00034-f. [DOI] [PubMed] [Google Scholar]
- 8.Walker MJ, Rylett CM, Keen JN, Audsley N, Sajid M, Shirras AD, Isaac RE. Proteome Sci. 2006;4:9. doi: 10.1186/1477-5956-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolfner MF, Harada HA, Bertram MJ, Stelick TJ, Kraus KW, Kalb JM, Lung YO, Neubaum DM, Park M, Tram U. Insect Biochem Mol Biol. 1997;27:825–834. doi: 10.1016/s0965-1748(97)00056-8. [DOI] [PubMed] [Google Scholar]
- 10.Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF. Proc Natl Acad Sci USA. 2001;98:7375–7379. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravi Ram K, Wolfner MF. Integr Comp Biol. 2007 Jun 1; doi: 10.1093/icb/icm046. [DOI] [PubMed] [Google Scholar]
- 12.Davies SJ, Chapman T. Insect Biochem Mol Biol. 2006;36:846–856. doi: 10.1016/j.ibmb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Andres JA, Maroja LS, Bogdanowicz SM, Swanson WJ, Harrison RG. Mol Biol Evol. 2006;23:1574–1584. doi: 10.1093/molbev/msl020. [DOI] [PubMed] [Google Scholar]
- 14.Collins AM, Caperna TJ, Williams V, Garrett WM, Evans JD. Insect Mol Biol. 2006;15:541–549. doi: 10.1111/j.1365-2583.2006.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryan JH. Nature. 1968;218:489. doi: 10.1038/218489a0. [DOI] [PubMed] [Google Scholar]
- 16.Bryan JH. Nature. 1972;239:519–520. doi: 10.1038/239519a0. [DOI] [PubMed] [Google Scholar]
- 17.Giglioli MEC, Mason GF. Proc R Entomol Soc London A. 1966;41:123–129. [Google Scholar]
- 18.Klowden MJ. J Insect Physiol. 2001;47:661–666. doi: 10.1016/s0022-1910(00)00127-x. [DOI] [PubMed] [Google Scholar]
- 19.Peng J, Zipperlen P, Kubli E. Curr Biol. 2005;15:1690–1694. doi: 10.1016/j.cub.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 20.Lung O, Tram U, Finnerty CM, Eipper-Mains MA, Kalb JM, Wolfner MF. Genetics. 2002;160:211–224. doi: 10.1093/genetics/160.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiumera AC, Dumont BL, Clark AG. Genetics. 2007;176:1245–1260. doi: 10.1534/genetics.106.064915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark AG, Aguade M, Prout T, Harshman LG, Langley CH. Genetics. 1995;139:189–201. doi: 10.1093/genetics/139.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravi Ram K, Ji S, Wolfner MF. Insect Biochem Mol Biol. 2005;35:1059–1071. doi: 10.1016/j.ibmb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Charlet M, Lagueux M, Reichhart JM, Hoffmann D, Braun A, Meister M. Eur J Biochem. 1996;241:699–706. doi: 10.1111/j.1432-1033.1996.00699.x. [DOI] [PubMed] [Google Scholar]
- 25.Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- 26.Roberts KP, Johnston DS, Nolan MA, Wooters JL, Waxmonsky NC, Piehl LB, Ensrud-Bowlin KM, Hamilton DW. Asian J Androl. 2007;9:508–514. doi: 10.1111/j.1745-7262.2007.00318.x. [DOI] [PubMed] [Google Scholar]
- 27.Ejima A, Smith BP, Lucas C, van der Goes van Naters W, Miller CJ, Carlson JR, Levine JD, Griffith LC. Curr Biol. 2007;17:599–605. doi: 10.1016/j.cub.2007.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller JL, Ripoll DR, Aquadro CF, Wolfner MF. Proc Natl Acad Sci USA. 2004;101:13542–13547. doi: 10.1073/pnas.0405579101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murer V, Spetz JF, Hengst U, Altrogge LM, de Agostini A, Monard D. Proc Natl Acad Sci USA. 2001;98:3029–3033. doi: 10.1073/pnas.051630698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danielli A, Kafatos FC, Loukeris TG. J Biol Chem. 2003;278:4184–4193. doi: 10.1074/jbc.M208187200. [DOI] [PubMed] [Google Scholar]
- 31.Meikle DB, Sheehan KB, Phillis DM, Richmond RC. J Insect Physiol. 1990;36:93–101. [Google Scholar]
- 32.Lung O, Kuo L, Wolfner MF. J Insect Physiol. 2001;47:617–622. doi: 10.1016/s0022-1910(00)00151-7. [DOI] [PubMed] [Google Scholar]
- 33.Samakovlis C, Kylsten P, Kimbrell DA, Engstrom A, Hultmark D. EMBO J. 1991;10:163–169. doi: 10.1002/j.1460-2075.1991.tb07932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asquith KL, Harman AJ, McLaughlin EA, Nixon B, Aitken RJ. Biol Reprod. 2005;72:328–337. doi: 10.1095/biolreprod.104.034470. [DOI] [PubMed] [Google Scholar]
- 35.Tutuncu L, Stein P, Ord TS, Jorgez CJ, Williams CJ. Dev Biol. 2004;270:246–260. doi: 10.1016/j.ydbio.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Zdobnov EM, von Mering C, Letunic I, Torrents D, Suyama M, Copley RR, Christophides GK, Thomasova D, Holt RA, Subramanian GM, et al. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- 37.Catteruccia F, Benton JP, Crisanti A. Nat Biotechnol. 2005;23:1414–1417. doi: 10.1038/nbt1152. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Tamura K, Nei M. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.