Abstract
Transcription factors play a key role in integrating and modulating biological information. In this study, we comprehensively measured the changing abundances of mRNAs over a time course of activation of human peripheral-blood-derived mononuclear cells (“macrophages”) with lipopolysaccharide. Global and dynamic analysis of transcription factors in response to a physiological stimulus has yet to be achieved in a human system, and our efforts significantly advanced this goal. We used multiple global high-throughput technologies for measuring mRNA levels, including massively parallel signature sequencing and GeneChip microarrays. We identified 92 of 1,288 known human transcription factors as having significantly measurable changes during our 24-h time course. At least 42 of these changes were previously unidentified in this system. Our data demonstrate that some transcription factors operate in a functional range below 10 transcripts per cell, whereas others operate in a range three orders of magnitude greater. The highly reproducible response of many mRNAs indicates feedback control. A broad range of activation kinetics was observed; thus, combinatorial regulation by small subsets of transcription factors would permit almost any timing input to cis-regulatory elements controlling gene transcription.
Keywords: gene expression microarray, massively parallel signature sequencing, systems biology, transcript enumeration
Systems biology has advanced our understanding of regulatory networks in unicellular and multicellular organisms (1–3). Developing methodologies for identifying and understanding the dynamics of the expression of all of the genes operating in a human system is a priority for systems biology. Human systems are exceptionally difficult because of their complexity and experimental constraints. A paradigm for understanding nuclear regulation is to (i) identify all transcription factors (TFs) involved in regulating a system, (ii) identify their targets based on computational predictions of binding to TF binding sites as well as perturbations in expression of target genes, and (iii) incorporate this knowledge into a predictive network model with genes as nodes and binding interactions as edges (4, 5). This approach works well when there is substantial preexisting knowledge enabling the execution of this paradigm. However, in humans, the majority of binding sites and targets for most TFs in most cell conditions are not known and cannot be predicted with confidence. Therefore, the development of a comprehensive model of nuclear regulation in most human tissues, including stimulated macrophages, remains beyond the reach of current research efforts. What is required now to achieve these comprehensive models is hard work. For most systems of interest, multiple studies will be required. These studies will require separate publication to ensure adequate documentation of the methodology and analysis, to allow separate groups to make distinct contributions, to allow different facets of a system to be explored in depth, to facilitate data deposition, and to ensure that dissemination of some results are not delayed while publications are held for other results to mature.
In this report, we describe one strategy for the comprehensive identification and dynamic analysis of the mRNA components in a system. In this model system, we activated peripheral-blood-derived mononuclear cells, which can be loosely termed “macrophages,” with lipopolysaccharide (LPS). We focused on the precise measurement of mRNA concentrations. There is currently no high-throughput technology that can precisely and sensitively measure all mRNAs in a system, although such technologies are likely to be available in the near future. To demonstrate the potential utility of such technologies, and to motivate their development and encourage their use, we produced data from a combination of two distinct current generation technologies and extensive hand curation that we believe will approximate the comprehensiveness and sensitivity of anticipated transcriptome technologies. An example of such a technology would be transcript enumeration based on unbiased sampling and sequencing of millions of transcripts (e.g., Solexa expression profiling; Illumina, San Diego, CA).
Macrophages provide a model for the study of mammalian signaling pathways. Many aspects of their behavior in cell culture are similar to their in vivo behavior. Toll-like receptors recognize ligands characteristic of pathogen activity such as LPS (6). The activation of macrophages by LPS through Toll-like receptor 4 is a model for perturbing regulatory networks in cells; our work is part of a general effort to study the nature of information transfer in mammalian intracellular networks (7, 8). Our data permit insight into the information encoded by TF mRNA concentrations that is subsequently transduced into other forms of information. Changes in mRNA concentration after a stimulus may indicate the importance of that transcript in two manners (9, 10): (i) the change in concentration may directly transfer information as part of the response pathway, and (ii) the change in concentration may be an indirect consequence of the response of a product of that transcript, such as to replenish a consumed protein.
We used two distinct high-throughput methodologies for dynamic mRNA analysis: a transcript enumeration methodology, massively parallel signature sequencing (MPSS), and a microarray hybridization methodology, GeneChips (Affymetrix, Santa Clara, CA) (11). Better coverage for gene identification is obtained because each technology has unique systematic errors, often not present in the other technology. MPSS permits the mean number of transcripts per cell to be approximated from transcripts per million (tpm) by knowing the approximate total number of transcripts per cell. Macrophages have ≈320,000 transcripts per cell (12).
The primary purpose of this report was to provide a building block for the systems biology paradigm of understanding macrophage nuclear regulation by providing a list of TFs that are almost certainly key nodes in intracellular information processing. Our aim was to capture all such TFs, but the following factors were missed: (i) TFs without changes in expression across our sampled time points and (ii) TFs not adequately assayed by the combination of technologies used. Despite these limitations, we almost doubled the number of TFs known to be involved in macrophage activation from 50 to 92. The core decision to be made for each TF is Boolean: to either recommend or not recommend inclusion of a TF in a predictive network model for macrophage function. We asserted that this decision in human systems is currently best done by using a combination of (i) automated algorithms for managing and analysis of high-throughput data and (ii) expert curation applied to each decision in the context of current accumulated knowledge of the system. The use of subjective expert analysis has been the standard for single-gene studies for many decades; it will be a long time, if ever, before such analysis is obsolete. Automated algorithms produce significance statistics and ranking for each gene for each technology and increasingly can usefully combine these statistics. Expert curation permits careful analysis of raw data and biological context for each gene. To derive maximum utility from the time available for expert curation, we chose to focus in this study exclusively on TFs. This choice is motivated by the important role TFs play in information transduction. For many genes, we anticipated an unprecedented sensitivity with our strategy, because the maximum sensitivity of either Affymetrix or MPSS technology alone could be estimated to be approximately one transcript per cell.
Results
List of Human TFs.
To identify important TFs, we first generated a comprehensive list of all TFs. The resulting list, selected from the 25,204 entries in Entrez Gene as of 2004, included 1,288 genes with enough evidence to be considered “known” TFs; the results are summarized online (as file Compilation_of_Mammalian_Transcription_Factors; www.systemsbiology-immunity.org). This list represents a snapshot of current knowledge (13). The importance to the community of a reference list of TFs is illustrated by the independent parallel development of such lists (14).
Multiple High-Throughput Datasets.
We isolated human peripheral-blood-derived monocytes and stimulated them with LPS over a 24-h time course (time points: basal, 2, 4, 8, and 24 h). We assayed TF expression with two independent technologies: MPSS and GeneChips (11, 15). Affymetrix data are available in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE5504); full annotated data are also available (as the file MPSS_data; www.systemsbiology-immunity.org/datadownload.php). Analysis of our combined data in the context of external information (e.g., tissue specific expression datasets) increased our ability to rank genes by biological significance. For reporting TFs, we selected a cutoff based on the false discovery rate that would have been present in the absence of expert curation of our raw data (see Materials and Methods); in short, we ranked the genes in order of increasing uncurated P values and then applied expert curation to each gene in the list until 10 consecutive genes were deemed not to belong. We sought to identify TFs with mRNA expression levels that have a measurably significant change between at least two time points during the LPS stimulation time course and, subsequently, to infer that these were likely involved in information transduction. We also observed TFs with a constant level of expression significantly above zero. In addition to the TFs that we identified as having significantly changing mRNA concentration, the TFs with constant mRNA level may also transduce information. Before our study, we compiled a list of TFs that could be considered to be known previously to be involved in macrophage activation; the results are summarized online (in subsection Biological Knowledge of Each TF of the file Ancillary_data of our online data; www.systemsbiology-immunity.org/datadownload.php). To summarize the online material, ≈50 TFs were known previously to be involved. In our present study, we identified all of these previously known TFs and then added ≈50, thus bringing the total to ≈100. Our statistical analysis suggests that few additional genes in this system will be found by further transcriptional analysis.
We confirmed the significance of change in mRNA levels for 15 TFs from our final list with real-time PCR (www.systemsbiology-immunity.org/datadownload.php): MYBL2, SPI1, MSC, STAT1, THRA, MXD1, HHEX, KLF16, MXI1, JUNB, IRF7, IRF3, AHR, ATF3, and ETS1. We selected IRF3, KLF16, HHEX, MXI1, and MYBL2 because they are at the limit of sensitivity of our technique. We selected KLF16, MSC, MXI1, and THRA because they had not been identified previously in this system. IRF7 and MSC were selected as positive controls because they are strongly and clearly regulated significantly at a high level. ATF3 was selected to evaluate changes at a moderate level. SPI1 and MXD1 were selected because the dynamic profiles in our MPSS and Affymetrix data differed. All 15 were confirmed as having a significantly measurable mRNA change. The likelihood of this result would be ≈95% if we had a false discovery rate of 0.1% and 50% if we had a false discovery rate of 4.5%. Our false-negative rate is hard to estimate, although we found all genes known in the literature to have expression changes in the LPS stimulation system in the time scale considered. False negatives would most likely result from biases present in both technologies. Currently, the major sources of error in high-throughput technologies lie within bioinformatics pipelines. Such errors include genes with improperly annotated polyadenylation sites and tags or array identifiers assigned to the wrong source.
Examples of Extreme Sensitivity.
The expression levels seen span from one transcript per cell to thousands of transcripts per cell. As measured by MPSS, seven significantly changing TFs had maximum expression of <12 tpm (approximately 4 transcripts per cell): KLF4, HHEX, MXI1, ETS1, HIVEP2, DBP, and CDCA7L. MPSS is known to underreport KLF4 tag counts because of a technology-specific bias (the combination of tetramers TTTA in the four-stepper and TCAA in the two-stepper frames); therefore, KLF4 is not an example of extreme sensitivity. HHEX and MXI1 both with a peak of 6 tpm (approximately two transcripts per cell) were the two cases of highest sensitivity in our analysis (and are not predicted to have underreported MPSS tetramer combinations). Our sensitivity is not universal and is less for some genes than others. Unrecognized biases in the technologies might account for some of these data.
Most TFs Are Not Transcriptionally Modulated in Macrophages.
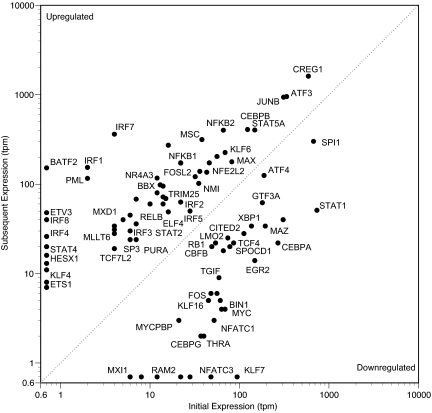
Evolutionary and complexity theory predicts that most (i.e., >50% and perhaps >90%) TFs are not involved in transducing information in any given specialized cellular state (16). This prediction rests on the hypothesis that many, but not all, TFs will have specific functions applicable to only a few cell types or conditions. When TFs with no expression were included (i.e., those that stayed at 0 tpm across the entire time course), there were 1,220 TFs with no significantly measurable MPSS change between any of our conditions. For example, the most highly expressed TF, YBX1, averaging 3,500 tpm, showed no significant change by our analysis. The next most abundant unchanging TF mRNAs were those of ZNF90, ZNF114, and EDF1, each with ≈450 tpm. The only TFs with higher average expression were CREG1, ATF3, JUNB, and SPI1, all with significant changes (Fig. 1).
Fig. 1.
Absolute expression levels of TFs that significantly change during macrophage activation. This graph is organized under the simplifying assumption that all TFs are either “induced” or “repressed.” To transform sometimes more complex expression time courses for this formalization, we retained only the lowest and highest expression level for each 24-h time course. If the lowest value preceded the highest, the TF was classified as induced; otherwise, the TF was classified as repressed. For some dynamics (e.g., oscillatory), this display of maxima and minima is a considerable simplification; Figs. 3 and 4 provide complete time courses. For visualization, values of 0 have been replaced with 0.7 before logarithmic transformation. One transcript per cell is ≈3 tpm. IFN response factors (IRFs) are well represented in the group of TFs that are induced from trace to high levels. Induced genes (above the dashed line) outnumber repressed genes (below the dashed line). To avoid clutter, not all points are labeled.
TFs Active in Macrophages.
Based on our analysis, a total of 92 TFs change significantly between at least two time points in our system; 43 of these have never been suspected to be involved in macrophage activation. Of these 92 TFs, 52 are induced, 31 are repressed, and 8 oscillated.
Timing of TF Expression.
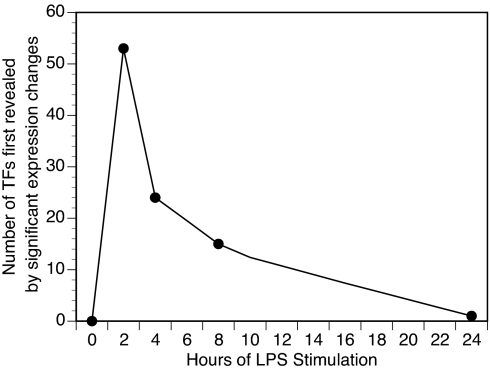
TF effects can be mediated and modulated by diverse molecular mechanisms. The quality and quantity of information transduced cannot always be inferred from examination of expression levels alone. In particular, the peak action of a TF can occur before the peak expression level. For example, the target gene of a TF might reach its maximal transcription rate when the concentration of that TF message is only 10% of its eventual maximum. This would occur if binding of a target promoter were saturated. Therefore, it is useful to classify TFs by the timing at which they first show a significant change. An advantage of this classification is that it reduces the dimensionality of the data (Fig. 2).
Fig. 2.
Most TFs that are going to change do so relatively early, indicating that much of the cellular regulatory logic occurs in the first few hours following stimulation. The origin is included arbitrarily for reference. This representation is intended to emphasize the spike in onset of TF expression change. However, many of the TFs that first show significant change at early time points remain significantly changed at later time points (Figs. 3 and 4).
Of the TFs that changed significantly, change tended to be apparent early. Most (52 of 92) showed significant change by 2 h (Fig. 3). There was a sharp drop in the number of TFs that first showed significant change at subsequent time points (Fig. 4). This early “spike” followed by little additional TF recruitment suggests that these macrophages were approaching a new state of TF expression by 24 h after stimulation (Fig. 2). Of the 92 TFs that showed a significant change during our time course, 50 TFs would have shown a significant change if only the basal and 24-h time points were considered. Thus, many of the changing TFs remained in their altered transcriptional state for at least 24 h.
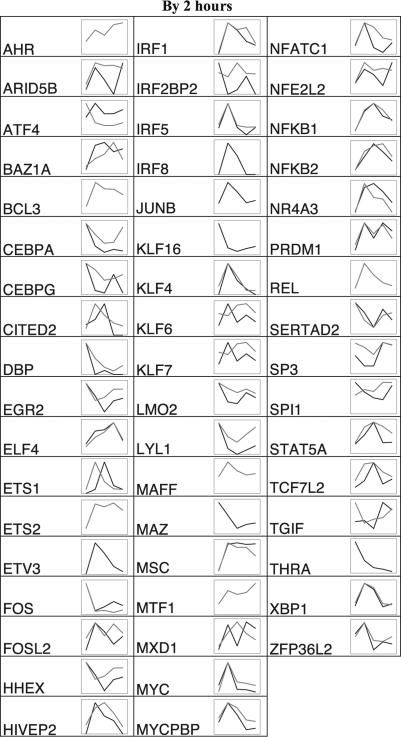
Fig. 3.
Shape of TF expression changes in stimulated macrophages with measurements normalized to the value of the highest time point. TFs showing a first significant time point at 2 h are shown. Black, MPSS tpm; gray, Affymetrix signal. Abscissa time points (not to scale) are basal and 2, 4, 8, and 24 h after addition of LPS. Raw values and statistics are recorded in the online material.
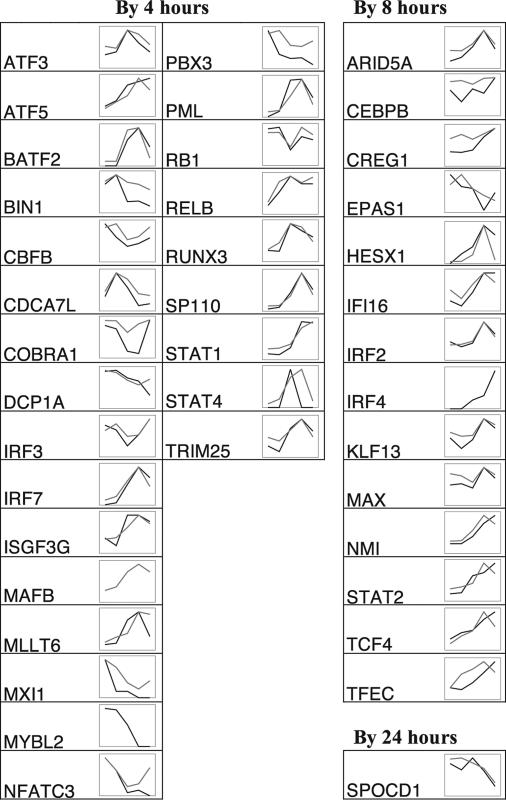
Fig. 4.
Shape of TF expression changes in stimulated macrophages with measurements normalized to the value of the highest time point. TFs showing a first significant time point of 4, 8, or 24 h are shown. Black, MPSS tpm; gray, Affymetrix signal. Abscissa time points (not to scale) are basal and 2, 4, 8, and 24 h after addition of LPS. Raw values and statistics are recorded in the online material.
The dynamic profiles of each TF can be classified by a few gross shapes: “peak,” “trough,” “up,” “down,” and “multiple inflections” (Figs. 3 and 4). Of those with straightforward classifications, there were 30 peak shapes, and 9 trough shapes, 23 were up, 17 were down, and 4 showed multiple inflections. Our goal in assigning dynamic shapes to TFs was to provide gross insight but not to define a formally rigorous classification; such a classification would require more time points to better exclude missed inflection points.
Consistency of Expression Data.
The normalized MPSS and Affymetrix profiles overlie each other almost exactly for 53 of the 92 TFs. The samples for each data set were independent biological samples from blood samples from different sets of unrelated individuals, acquired and processed months apart. Therefore, the concordance of many of the profiles, derived from different technologies, technical replicates, and biological samples, is striking and suggests that some of these genes are under tight regulation not only as to the timing of their expression peak but also as to their exact expression level at each time point. Such genes include ELF4, KLF4, IRF2, MYC, NFKB1, XBP1, GTF2B, and ARID5A. The remaining 39 profiles are either defined by data from only one of the two technologies or are discordant between the two technologies (Figs. 3 and 4).
Discussion
Previously ≈49 TFs were known to be involved in macrophage signaling. This estimate is based on very generous criteria: the actual number found by a macrophage expert performing a literature search would be less; we inferred inclusion for some TFs in this list from previously existing online array datasets. We have approximately doubled the number of previously identified TFs in the macrophage response to LPS to ≈100. We use the “approximate” qualifier (i) because this figure depends on whether or not cofactors and basal TFs are included, (ii) to account for uncertainties in the definition of TF and cofactor, and (iii) to allow for statistical uncertainty in the data analysis. We manually examined the data in the context of legacy data and the literature for all 1,288 known TF genes (≈5% of all known genes) with the exception of genes for which all transcript variants (i) showed no significant change in either MPSS or Affymetrix assays across our time course, (ii) had no prior expectation from the literature of being involved in macrophage activation, and (iii) did not have an average expression level exceeding 50 tpm in our MPSS data. This process resulted in a Boolean decision to include the TF in our list of genes relevant to the systems biology of macrophage activation. There is no single P value associated with each of these decisions. Rather, each decision is based potentially on disparate data from different sources and is justified by a discussion specific to each gene (www.systemsbiology-immunity.org/datadownload.php), much as would be found in the discussion section of a journal article that focused on a single gene. In this context, the most important criteria were the set of P values for the significance of changes in mRNA concentrations between pairs of conditions as measured by our two technologies.
A “Parts List” for Systems Biology.
Significant differences in expression measurements between two cellular states imply that a TF is an important part of the regulatory network driving or effectuating that state transition. However, significant expression changes are not required for significant information transduction; many TFs effectuate state changes in cells through changes in protein modification or localization (e.g., steroid hormone receptors). Before our study, 49 TFs were known to have significant mRNA changes; we confirmed these data (Fig. 1; notes provided by the curator for each gene are archived at www.innateimmunity-systemsbiology.org). All TFs with known expression changes in macrophage activation in the literature were found by our analysis except for those that could not be detected by our experimental design. For example, EGR1, ID2, JUN, and SP2, which peak before our earliest time point at 2 h went undetected by our technique, as did RELA, which does not have significant mRNA expression changes.
Our work offers a comprehensive view of all TFs operating in a mammalian system responding to a perturbing signal. Therefore, it may offer insights general to the understanding of nuclear regulation in many mammalian systems. We have elucidated the number of TFs operating in a system. This number, 92, is likely to be a lower bound because we strove to avoid false positives at the possible cost of an increased number of false negatives, most of which were likely due to genes that were currently unknown or not annotated as TFs. We estimate an upper bound to be considerably less than 200 because we believe current annotation gaps plus our technological false-negative rate would not inflate our number beyond this limit. For the purposes of text mining, the alphabetical list of 92 TFs determined by expression analysis to be important to information transduction in macrophages is: AHR, ARID5A, ARID5B, ATF3, ATF4, ATF5, BATF2, BAZ1A, BCL3, BIN1, CBFB, CDCA7L, CEBPA, CEBPB, CEBPG, CITED2, COBRA1, CREG1, DBP, DCP1A, EGR2, ELF4, EPAS1, ETS1, ETS2, ETV3, FOS, FOSL2, HESX1, HHEX, HIVEP2, IFI16, IRF1, IRF2, IRF2BP2, IRF3, IRF4, IRF5, IRF7, IRF8, ISGF3G, JUNB, KLF13, KLF16, KLF4, KLF6, KLF7, LMO2, LYL1, MAFB, MAFF, MAX, MAZ, MLLT6, MSC, MTF1, MXD1, MXI1, MYBL2, MYC, MYCPBP, NFATC1, NFATC3, NFE2L2, NFKB1, NFKB2, NMI, NR4A3, PBX3, PML, PRDM1, RB1, REL, RELB, RUNX3, SERTAD2, SP110, SP3, SPI1, SPOCD1, STAT1, STAT2, STAT4, STAT5A, TCF4, TCF7L2, TFEC, TGIF, THRA, TRIM25, XBP1, ZFP36L2. For the purpose of text mining, a simplified list of TFs identified primarily from analysis of Affymetrix data are: AHR, BCL3, ETS2, MAFB, MAFF, MTF1, REL. For the purpose of text mining, a simplified list of TFs identified primarily from analysis of MPSS data are: ETV3, IRF4, IRF8, JUNB, KLF16, MAZ, MYBL2, THRA.
Within 2 h of LPS stimulation, our results demonstrate that the nuclear regulation response in macrophages became very complex. Most TFs that were going to change their transcription at any point in the activation process had already done so by 2 h. The number of never-before-recruited TFs dropped considerably with each time point. By 24 h, almost no new TFs were being recruited. We predict that the 50 TFs that sustain their significant change (either up or down) would include those necessary for maintaining differentiated states such as LPS tolerance, whereas the other TFs (typically showing a peak or trough) would be responsible primarily for either converting to a differentiated state or effectuating transient responses such as cytokine release. Many of the active TFs in the system were under exquisite control. They showed the same relative amplitude and timing of transcriptional response in distinct biological samples, drawn from separate individuals years apart, measured with distinct technologies. These genes are most likely under the influence of multiple levels of feedback regulation that are robust to environmental and allelic variation. The concentrations of the active TFs span several orders of magnitudes. There is no clear prototypical operational range for the concentration of a TF. Some TFs exert influence by changing their concentration from zero to a few transcripts per cell, whereas others alter their concentration between levels of hundreds to thousands of transcripts per cell. The dynamic profiles (mRNA concentrations as a function of time) of the TFs are diverse. Profiles have distinct times for maxima and minima and relative ordering of these points. This implies that if a cell were to use these as inputs to a signal generator, it could produce almost any signal by mixing them combinatorially.
A recent study incorporated mouse macrophage TFs in network analysis (17, 18). The authors chose four TFs to discuss: ATF3, ETS2, IRF1, and NFE2L2 (also known as NRF2). All four of these were among the 92 identified by our study. In the study we report here, we chose to defer inferring a network, much as one might defer assembling a jigsaw puzzle missing many of its pieces. Although we may have identified most of the key TF nodes, little is known about these nodes. Interactions of most of these TFs cannot be reliably inferred. An urgent need now is to experimentally determine binding sites of the majority of TFs transducing information in macrophages. Approaches should include experimental determination of binding-site matrices (enabling computational approaches) and direct determination of TF target genes with techniques such as chromatin immunoprecipitation. A primary result and conclusion of our study is the identification of the need for these future experiments and of the TFs that should be the primary subjects of these experiments.
We specifically embarked on one such follow-up study. At the beginning of this project, ATF3 was included in the list of genes known to be involved in macrophage activation (19), but only 10 target genes were known. Only three of these are expressed by macrophages: ATF3 (autoregulation), ASNS, and MMP2. None of these three interactions was present in curated databases; each had to be inferred from literature searches. Therefore, from a systems perspective, there was essentially no prior knowledge of ATF3 in relation to macrophage activation. This was and remains true for most of the TFs that we identified in the present work. From analysis of the dynamic profile of ATF3 in this study coupled with a parallel study in a murine model system, we developed a hypothesis that ATF3 might be an important global regulator of macrophage activation. Its high absolute level of expression (Fig. 1) was suggestive that it might be a pervasive global regulator that operated on many genes, with a weak affinity for promoter elements necessitating not only the high level of expression but also cooperative regulation with other TFs. As an aside, we also hypothesized that TFs with low levels of expression such as KLF4 and HESX1 would also play important roles but were more likely to bind more strongly to a more restricted set of target genes. For ATF3, hypothesis-driven follow-up experiments in the mouse involved fine-scale time courses, additional stimuli, genetic perturbations, and chip-to-chip TF–DNA binding analysis. They resulted in confirmation of the important cooperative role of ATF3 in macrophage activation and expanded the list of ATF3 targets (19).
We have achieved one milestone along the path to the realization of systems biology in humans. We obtained a comprehensive overview of the dynamics of expressed transcripts after stimulation of macrophages with LPS. Combining the strengths of two complementary high-throughput technologies, our analysis reached a sensitivity of close to one transcript per cell with a dynamic range spanning five orders of magnitude. Our approach is applicable to other mammalian cells and tissues.
Materials and Methods
Curation of List of TFs.
Information in Entrez Gene, Online Mendelian Inheritance in Man, PubMed, GeneOntology, coupled with PFAM protein domain content analysis with HMMER was analyzed to produce a list of human TFs. Inclusion was necessarily subjective for two reasons: (i) the definition of TF is imprecise, and (ii) there is not enough information available for many genes to be certain of their function. Selection was guided by the following definition of TF: “a protein that is part of a complex at the time that complex binds to DNA with the effect of modifying transcription.” We included cofactors in our definition. To be considered, a gene had to have an Entrez Gene entry. The list of TFs selected from the 25,204 entries in Entrez Gene with assigned sequences includes 1,288 genes with enough evidence to be considered “known” TFs; of these, 564 have a maximum expression in our data of at least 6 tpm, and thus could be considered a candidate list of TFs present in macrophages. A value of 6 tpm represents approximately three transcripts per cell. We use the cutoff of 6 tpm because it is the maximum expression level of HHEX, which has the smallest maximum expression level of all of the TFs that we determined to have significantly changing expression levels.
Cells and RNA.
Adherent monocytes were isolated from peripheral blood mononuclear cells collected from five healthy humans for MPSS studies and three healthy humans (distinct from the MPSS donors) for the Affymetrix studies and cultured for 10 days in RPMI medium 1640 (20% FBS/l-glutamine/20 mM Hepes/penicillin/streptomycin/50 ng/ml macrophage colony-stimulating factor) to generate peripheral-blood-derived mononuclear cells. Peripheral-blood-derived mononuclear cells were stimulated with 100 ng/ml LPS (Salmonella minnesota R595 ultrapure LPS; List Biological Laboratories, Campbell, CA) and sampled at time points 0 (i.e., before stimulation), 2, 4, 8, and 24 h. For each of these time points, total RNA was isolated with TRIzol (Invitrogen, Carlsbad, CA). Because of the expense of MPSS, but to maintain our ability to detect significant changes between conditions, RNA from the five donors was pooled for each time point. This was done in duplicate sets of five for the basal and 4-h time points and a single set for the other time points. PolyA RNA was isolated with a MicroPoly(A-)Pure kit (Ambion, Austin, TX). Supernatants were tested to confirm appropriate induction of cytokines (TNF, IL-6, and IL-12), and an aliquot of total RNA was tested by using real-time PCR to ensure appropriate induction of selected genes. For Affymetrix samples, the macrophages from each of the three donors were stimulated with 100 ng/ml LPS for 0, 2, 4, 8, or 24 h. For each of these samples and time points, total RNA was isolated with TRIzol and probes were prepared by using the Affymetrix protocol. We estimated 320,000 transcripts per cell: the average yield was 1.58 μg of polyA mRNA from five million cells per preparation, assuming an 1800-bp average transcript length. Analysis of our MPSS data verified that transcripts specific for nonmacrophage blood cells were not present in the RNA.
Signature Cloning and MPSS.
Signature Cloning and MPSS were performed according to standard protocols (11). Raw data were reported as tpm for each bead that produced at least 17 bp of sequence. Initially, all mRNA and EST sequences annotated to an Entrez Gene in the National Center for Biotechnology Information database were scanned for all possible GATC sites, retaining in the pipeline those signatures most likely to appear in MPSS analysis (e.g., those immediately 5′ to a predicted polyA). The impact of polymorphism was largely absorbed by the use of UniGene data for each National Center for Biotechnology Information (NCBI) gene identified (20). Further manual curation was performed as necessary.
Affymetrix.
mRNA was prepared and two samples for each condition were analyzed on HG-U133 Plus2 GeneChips.
Statistics.
The tpm for genes was calculated as the sum of a set of signatures representing that gene. This set may contain signatures representing different gene products. However, the P value assigned to a gene is determined only from a single signature: that with the highest mean tpm (almost always the most 3′ signature in a gene). MPSS statistics were computed as described (21). Affymetrix statistical analysis was per Ideker et al. (22). In an initial screening, genes were ranked by the most significant P value produced by either methodology. The most significant and all TFs with disagreements between the methodologies were examined by expert curation (in subsection “Supplemental Comments on the Evaluation of Genes” of the file Ancillary_data of our online data; www.innateimmunity-systemsbiology.org). This examination included consideration of Affymetrix probe-level data, manual mapping of tags to genes, time-course profile, signal-to-noise ratios, tissue expression profile (23), and the literature. The inclusion of a given TF might be made solely on the basis of exceptionally strong evidence in the literature even in the absence of a significant P value for a particular technology. Alternatively, inclusion of a given gene might be made solely on the basis of our newly acquired data. The sensitivity and specificity of our strategy varies across genes and depends on many factors, including the sequence of the MPSS tags, the efficiency of hybridization of the Affymetrix ProbeSet, the quality of the bioinformatics mappings for information associated with the gene, and the knowledge in legacy databases and the literature. A threshold for choosing the genes to report in Fig. 1 was set as the rank at which 10 consecutive TFs sorted by increasing P value were deemed not to belong following curation. This threshold was determined by the perceived marginal value of allocating additional hours of expert curation for decreasing return.
Real-Time PCR.
TFs were selected for verification from the high end of the dynamic range (e.g., JUNB), the low end of the dynamic range (e.g., HHEX), and from those that were discrepant between MPSS and Affymetrix measurements (e.g., SPI1). Assays were read on an MX3005P system (Stratagene, La Jolla, CA).
Acknowledgments
This study received support from National Institute of Allergy and Infectious Diseases/National Institutes of Health Grants 5K08AI056092 and 5U54AI054523.
Abbreviations
- LPS
lipopolysaccharide
- MPSS
massively parallel signature sequencing
- TF
transcription factor
- tpm
transcripts per million.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Affymetrix data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE5504).
References
- 1.Baliga NS, Bjork SJ, Bonneau R, Pan M, Iloanusi C, Kottemann MC, Hood L, DiRuggiero J. Genome Res. 2004;14:1025–1035. doi: 10.1101/gr.1993504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papin JA, Hunter T, Palsson BO, Subramaniam S. Nat Rev Mol Cell Biol. 2005;6:99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- 3.Davidson EH, McClay DR, Hood L. Proc Natl Acad Sci USA. 2003;100:1475–1480. doi: 10.1073/pnas.0437746100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Atauri P, Sorribas A, Cascante M. Biotechnol Bioeng. 2000;68:18–30. doi: 10.1002/(sici)1097-0290(20000405)68:1<18::aid-bit3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 5.Zak DE, Pearson RK, Vadigepalli R, Gonye GE, Schwaber JS, Doyle FJ., III Omics. 2003;7:373–386. doi: 10.1089/153623103322637689. [DOI] [PubMed] [Google Scholar]
- 6.Takeda K, Kaisho T, Akira S. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 7.Ideker T, Galitski T, Hood L. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 8.Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A. Proc Natl Acad Sci USA. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chadalavada RS, Korkola JE, Houldsworth J, Olshen AB, Bosl GJ, Studer L, Chaganti RS. Stem Cells. 2007;25:771–778. doi: 10.1634/stemcells.2006-0271. [DOI] [PubMed] [Google Scholar]
- 10.Gaudet S, Janes KA, Albeck JG, Pace EA, Lauffenburger DA, Sorger PK. Mol Cell Proteomics. 2005;4:1569–1590. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Zhou D, Rao MS, Walker R, Khrebtukova I, Haudenschild CD, Miura T, Decola S, Vermaas E, Moon K, Vasicek TJ. Methods Mol Biol. 2006;331:285–311. doi: 10.1385/1-59745-046-4:285. [DOI] [PubMed] [Google Scholar]
- 12.Glen AC. Clin Chem. 1967;13:299–313. [PubMed] [Google Scholar]
- 13.Glusman G, Qin S, El-Gewely MR, Siegel AF, Roach JC, Hood L, Smit AF. PLoS Comput Biol. 2006;2:e18. doi: 10.1371/journal.pcbi.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong YM, Macdonald RJ, Wen X, Yang P, Barbera VM, Swift GH. Gene Expr Patterns. 2006;6:678–686. doi: 10.1016/j.modgep.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 16.Mattick JS, Gagen MJ. Mol Biol Evol. 2001;18:1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson R, Bajic VB, Suzuki H, di Bernardo D, Bjorkegren J, Katayama S, Reid JF, Sweet MJ, Gariboldi M, Carninci P, et al. Genomics. 2006;88:133–142. doi: 10.1016/j.ygeno.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 18.Tegnér J, Nilsson R, Bajic VB, Björkegren J, Ravasi T. Cell Immunol. 2006;244:105–109. doi: 10.1016/j.cellimm.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Kennedy K, Hai T, Bolouri H, Aderem A. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 20.Silva AP, De Souza JE, Galante PA, Riggins GJ, De Souza SJ, Camargo AA. Nucleic Acids Res. 2004;32:6104–6110. doi: 10.1093/nar/gkh937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stolovitzky GA, Kundaje A, Held GA, Duggar KH, Haudenschild CD, Zhou D, Vasicek TJ, Smith KD, Aderem A, Roach JC. Proc Natl Acad Sci USA. 2005;102:1402–1407. doi: 10.1073/pnas.0406555102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ideker T, Thorsson V, Siegel AF, Hood LE. J Comput Biol. 2000;7:805–817. doi: 10.1089/10665270050514945. [DOI] [PubMed] [Google Scholar]
- 23.Jongeneel CV, Delorenzi M, Iseli C, Zhou D, Haudenschild CD, Khrebtukova I, Kuznetsov D, Stevenson BJ, Strausberg RL, Simpson AJ, et al. Genome Res. 2005;15:1007–1014. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]