Abstract
A comprehensive analysis of both the molecular genetic and phenotypic responses of any organism to the space flight environment has never been accomplished because of significant technological and logistical hurdles. Moreover, the effects of space flight on microbial pathogenicity and associated infectious disease risks have not been studied. The bacterial pathogen Salmonella typhimurium was grown aboard Space Shuttle mission STS-115 and compared with identical ground control cultures. Global microarray and proteomic analyses revealed that 167 transcripts and 73 proteins changed expression with the conserved RNA-binding protein Hfq identified as a likely global regulator involved in the response to this environment. Hfq involvement was confirmed with a ground-based microgravity culture model. Space flight samples exhibited enhanced virulence in a murine infection model and extracellular matrix accumulation consistent with a biofilm. Strategies to target Hfq and related regulators could potentially decrease infectious disease risks during space flight missions and provide novel therapeutic options on Earth.
Keywords: microgravity, Space Shuttle, low shear modeled microgravity, rotating wall vessel, Salmonella
Environmental conditions and crew member immune dysfunction associated with space flight may increase the risk of infectious disease during a long-duration mission (1–4). However, our knowledge of microbial changes in response to space flight conditions and the corresponding changes to infectious disease risk is limited and unclear. Elucidation of such risks and the mechanisms behind any space flight-induced changes to microbial pathogens holds the potential to decrease risk for human exploration of space and provide insight into how pathogens cause infections in Earth-based environments. Numerous logistical and technological hurdles exist when performing biological space flight experimentation, and an extremely limited number of opportunities to perform such research are available. Accordingly, comprehensive analysis of cells, including pathogenic microbes, at the molecular and phenotypic level during space flight offers a rare opportunity to examine their behavior and response in this environment.
Previous studies using the enteric bacterial pathogen Salmonella enterica serovar Typhimurium showed that growth in a ground-based space flight analog bioreactor, termed the rotating wall vessel (RWV), induced global genotypic and phenotypic changes in this organism (5–7). Specifically, S. typhimurium grown in this space flight analog culture environment, described as low-shear modeled microgravity (LSMMG), exhibited increased virulence, increased resistance to environmental stresses (acid, osmotic, and thermal), increased survival in macrophages, and global changes in gene expression at the transcriptional and translational levels (5–7). Collectively, these results suggested the potential that the true space flight environment could globally alter bacterial genotypic and phenotypic responses. Thus, we designed an experimental approach to test our hypothesis, specifically to culture S. typhimurium during space flight and evaluate changes in microbial gene expression and virulence in response to this environment.
Our experiments were flown on Space Shuttle Atlantis Mission STS-115 (September 2006). In this experiment, cultures of S. typhimurium were activated to grow in space for a specific time period and then either fixed in an RNA/protein fixative or supplemented with additional growth media after this time period [supporting information (SI) Fig. 3]. At 2.5 h after landing at Kennedy Space Center, the culture samples were recovered and subsequently used for whole-genome transcriptional microarray and proteomic analysis (fixed samples) or for infections in a murine model of salmonellosis (media-supplemented samples). In each case, the flight culture samples were compared with culture samples grown under identical conditions on the ground at Kennedy Space Center using coordinated activation and termination times (by means of real-time communications with the Shuttle crew) in an insulated room that maintained temperature and humidity identical to those on the Shuttle (Orbital Environment Simulator). The culture experiments were loaded into specially designed hardware [termed fluid processing apparatus (FPA)] to facilitate controlled activation and fixation of the cultures while maintaining suitable culture containment requirements (SI Fig. 4).
To our knowledge, our results are the first documentation of changes in bacterial gene expression and microbial virulence in response to culture during space flight. Specifically, these findings demonstrate that the space flight environment imparts a signal that can induce molecular changes in bacterial cells. Furthermore, these results also provide direct evidence that this signal can alter the virulence of a microbial pathogen. Our collective data indicate that the conserved RNA-binding protein Hfq plays a central regulatory role in the microbial response to space flight conditions. Evaluation of microbial changes in response to this unique environment has the potential to provide heretofore unavailable insight into microbial response mechanisms to Earth-based environments, including those encountered by pathogens during the natural course of infection.
Results
Whole-Genome Transcriptional and Proteomic Analysis of Space Flight and Ground Cultures.
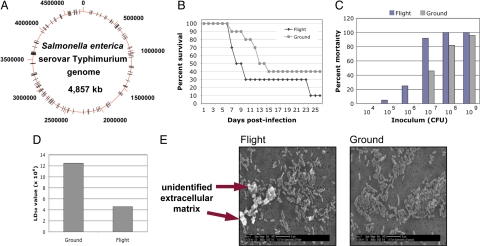
To determine which genes changed expression in response to space flight, total bacterial RNA was isolated from the fixed flight and ground samples, qualitatively analyzed to ensure lack of degradation by means of denaturing gel electrophoresis, quantitated, and then reverse-transcribed into labeled, single-stranded cDNA. The labeled cDNA was cohybridized with differentially labeled S. typhimurium genomic DNA to whole-genome S. typhimurium microarray slides. The cDNA signal hybridizing to each gene spot was quantitated, and the normalized, background-subtracted data were analyzed for statistically significant 2-fold or greater differences in gene expression between the flight and ground samples. We found 167 genes differentially expressed in flight as compared with ground controls from a variety of functional categories (69 up-regulated and 98 down-regulated) (SI Table 2). The proteomes of fixed cultures were also obtained by means of multidimensional protein identification analysis. We identified 251 proteins expressed in the flight and ground cultures, with 73 being present at different levels in these samples (SI Table 3). Several of the genes encoding these proteins were also identified by means of microarray analysis. Collectively, these gene expression changes form the first documented bacterial space flight stimulon indicating that bacteria respond to this environment with widespread alterations of expression of genes distributed globally throughout the chromosome (Fig. 1A).
Fig. 1.
Data from STS-115 S. typhimurium experiments. (A) Map of the 4.8-Mb circular S. typhimurium genome with the locations of the genes belonging to the space flight transcriptional stimulon indicated as black hash marks. (B) Decreased time to death in mice infected with flight S. typhimurium as compared with identical ground controls. Female BALB/c mice per-orally infected with 107 bacteria from either space flight or ground cultures were monitored every 6–12 h over a 30-day period, and the percent survival of the mice in each group is graphed versus the number of days. (C) Increased percent mortality of mice infected with space flight cultures across a range of infection dosages. Groups of mice were infected with increasing dosages of bacteria from space flight and ground cultures and monitored for survival over 30 days. The percent mortality (calculated as in ref. 23) of each dosage group is graphed versus the dosage amount. (D) Decreased LD50 value (calculated as in ref. 23) for space flight bacteria in a murine infection model. (E) SEM of space flight and ground S. typhimurium bacteria showing the formation of an extracellular matrix and associated cellular aggregation of space flight cells. (Magnification: ×3,500.)
Involvement of Hfq in Space Flight and LSMMG Responses.
Identification of one or more regulators of the space flight stimulon represents an important step in understanding the nature of this unique environmental signal. Our data indicated that a pathway involving the conserved RNA-binding regulatory protein Hfq played a role in this response (Table 1). Hfq is an RNA chaperone that binds to small regulatory RNA and mRNA molecules to facilitate mRNA translational regulation in response to envelope stress (in conjunction with the specialized σ factor RpoE), environmental stress (by means of alteration of RpoS expression), and changes in metabolite concentrations, such as iron levels (via the Fur pathway) (8–12). Hfq is also involved in promoting the virulence of several pathogens including S. typhimurium (13), and Hfq homologues are highly conserved across species of prokaryotes and eukaryotes (14). Our data strongly support a role for Hfq in the response to space flight: (i) The expression of hfq was decreased in flight, and this finding matched previous results in which S. typhimurium hfq gene expression was decreased in a ground-based model of microgravity (7). (ii) Expression of 64 genes in the Hfq regulon was altered in flight (32% of the total genes identified), and the directions of differential changes of major classes of these genes matched predictions associated with decreased hfq expression (see subsequent examples). (iii) Several small regulatory RNAs that interact with Hfq were differentially regulated in flight as would be predicted if small RNA/Hfq pathways are involved in a space flight response. (iv) The levels of OmpA, OmpC, and OmpD mRNA and protein are classic indicators of the RpoE-mediated periplasmic stress response, which involves Hfq (15). Transcripts encoding OmpA, OmpC, and OmpD (and OmpC protein level) were up-regulated in flight, correlating with hfq down-regulation. (v) Hfq promotes expression of a large class of ribosomal structural protein genes (12), and we found that many such genes exhibited decreased expression in flight. (vi) Hfq is a negative regulator of the large tra operon encoding the F plasmid transfer apparatus (16), and several tra genes from related operons on two plasmids present in S. typhimurium χ3339 were up-regulated in flight. (vii) Hfq is intimately involved in a periplasmic stress signaling pathway that depends on the activity levels of three key proteins, RpoE, DksA, and RseB; differential expression of these genes was observed in flight (8, 12). (viii) Hfq regulates the expression of the Fur protein and other genes involved in the iron response pathway, and we observed several iron utilization/storage genes with altered expression in flight (9, 11). This finding also matched previous results in which iron pathway genes in S. typhimurium changed expression in a ground-based model of microgravity, and the Fur protein was shown to play a role in stress resistance alterations induced in the same model (7).
Table 1.
Space flight stimulon genes belonging to Hfq regulon or involved with iron utilization or biofilm formation
| Gene | Fold change | Function |
|---|---|---|
| Hfq regulon genes (up-regulated) | ||
| Outer membrane proteins | ||
| ompA | 2.05 | Outer membrane porin |
| ompC | 2.44 | Outer membrane porin |
| ompD | 3.34 | Outer membrane porin |
| Plasmid transfer apparatus | ||
| traB | 4.71 | Conjugative transfer |
| traN | 4.24 | Conjugative transfer |
| trbA | 3.14 | Conjugative transfer |
| traK | 2.91 | Conjugative transfer |
| traD | 2.87 | Conjugative transfer |
| trbC | 2.68 | Conjugative transfer |
| traH | 2.59 | Conjugative transfer |
| traX | 2.37 | Conjugative transfer |
| traT | 2.34 | Conjugative transfer |
| trbB | 2.32 | Conjugative transfer |
| traG | 2.21 | Conjugative transfer |
| traF | 2.11 | Conjugative transfer |
| traR | 1.79 | Conjugative transfer |
| Various cellular functions | ||
| gapA | 7.67 | Glyceraldehyde dehydrogenase |
| sipC | 6.27 | Cell invasion protein |
| adhE | 4.75 | Fe-dependent dehydrogenase |
| glpQ | 2.58 | Glycerophosphodiesterase |
| fliC | 2.11 | Flagellin, structural protein |
| sbmA | 1.67 | ABC superfamily transporter |
| Hfq regulon genes (down-regulated) | ||
| Small RNAs | ||
| αRBS | 0.305 | Small RNA |
| rnaseP | 0.306 | Small RNA regulatory |
| csrB | 0.318 | Small RNA regulatory |
| tke1 | 0.427 | Small RNA |
| oxyS | 0.432 | Small RNA regulatory |
| RFN | 0.458 | Small RNA |
| rne5 | 0.499 | Small RNA |
| Ribosomal proteins | ||
| rpsL | 0.251 | 30S ribosomal subunit protein S12 |
| rpsS | 0.289 | 30S ribosomal subunit protein S19 |
| rplD | 0.393 | 50S ribosomal subunit protein L4 |
| rpsF | 0.401 | 30S ribosomal subunit protein S6 |
| rplP | 0.422 | 50S ribosomal subunit protein L16 |
| rplA | 0.423 | 50S ribosomal subunit protein L1 |
| rpme2 | 0.473 | 50S ribosomal protein L31 |
| rplY | 0.551 | 50S ribosomal subunit protein L25 |
| Various cellular functions | ||
| ynaF | 0.201 | Putative universal stress protein |
| ygfE | 0.248 | Putative cytoplasmic protein |
| dps | 0.273 | Stress response protein |
| hfq | 0.298 | Host factor for phage replication |
| osmY | 0.318 | Hyperosmotically inducible protein |
| mysB | 0.341 | Suppresses protein export mutants |
| rpoE | 0.403 | σE (σ24) factor |
| cspD | 0.421 | Similar to CspA; not cold-induced |
| nlpb | 0.435 | Lipoprotein-34 |
| ygaC | 0.451 | Putative cytoplasmic protein |
| ygaM | 0.453 | Putative inner membrane protein |
| gltI | 0.479 | ABC glutamate/aspartate transporter |
| ppiB | 0.482 | Peptidyl-prolyl isomerase B |
| atpE | 0.482 | Membrane-bound ATP synthase |
| yfiA | 0.482 | Ribosome-associated factor |
| trxA | 0.493 | Thioredoxin 1, redox factor |
| nifU | 0.496 | Fe-S cluster formation protein |
| rbfA | 0.506 | Ribosome-binding factor |
| rseB | 0.514 | Anti-σE factor |
| yiaG | 0.528 | Putative transcriptional regulator |
| ompX | 0.547 | Outer membrane protein |
| rnpA | 0.554 | RNase P, protein component |
| hns | 0.554 | DNA-binding protein |
| lamB | 0.566 | Phage λ receptor protein |
| rmf | 0.566 | Ribosome modulation factor |
| tpx | 0.566 | Thiol peroxidase |
| priB | 0.571 | Primosomal replication protein N |
| Iron utilization/storage genes | ||
| adhE | 4.76 | Fe-dependent dehydrogenase |
| entE | 2.24 | 2,3-dihydroxybenzoate-AMP ligase |
| hydN | 2.03 | Electron transport (FeS center) |
| dmsC | 0.497 | Anaerobic DMSO reductase |
| nifU | 0.495 | Fe-S cluster formation protein |
| fnr | 0.494 | Transcriptional regulator, Fe-binding |
| fdnH | 0.458 | Fe-S formate dehydrogenase-N |
| frdC | 0.411 | Fumarate reductase, anaerobic |
| bfr | 0.404 | Bacterioferrin, iron storage |
| ompW | 0.276 | Outer membrane protein W |
| dps | 0.273 | Stress response protein and ferritin |
| Genes implicated in/associated with biofilm formation | ||
| wza | 2.30 | Polysaccharide export protein |
| wcaI | 2.07 | Putative glycosyl transferase |
| ompA | 2.06 | Outer membrane protein |
| wcaD | 1.82 | Putative colanic acid polymerase |
| wcaH | 1.76 | GDP-mannose mannosyl hydrolase |
| manC | 1.71 | Mannose guanylyltransferase |
| wcaG | 1.68 | Bifunctional GDP fucose synthetase |
| wcaB | 1.64 | Putative acyl transferase |
| fimH | 1.61 | Fimbrial subunit |
| fliS | 0.339 | Flagellar biosynthesis |
| flgM | 0.343 | Flagellar biosynthesis |
| flhD | 0.356 | Flagellar biosynthesis |
| fliE | 0.438 | Flagellar biosynthesis |
| fliT | 0.444 | Flagellar biosynthesis |
| cheY | 0.461 | Chemotaxic response |
| cheZ | 0.535 | Chemotaxic response |
For specific parameters used to identify these genes, please see Materials and Methods.
Given these findings, we designed experiments to verify a role for Hfq in the space flight response using a cellular growth apparatus that serves as a ground-based model of microgravity conditions termed the RWV bioreactor (SI Fig. 5). Designed by the National Aeronautics and Space Administration, the RWV has been extensively used in this capacity to study the effects of a biomedically relevant low-fluid-shear growth environment (which closely models the liquid growth environment encountered by cells in the microgravity environment of space flight as well as by pathogens during infection of the host) on various types of cells (6, 17–19). Studies with the RWV involve using two separate apparatuses: one is operated in the low shear modeled microgravity position (LSMMG), and one is operated as a control in a position (termed 1 × g) where sedimentation due to gravity is not offset by the rotating action of the vessel.
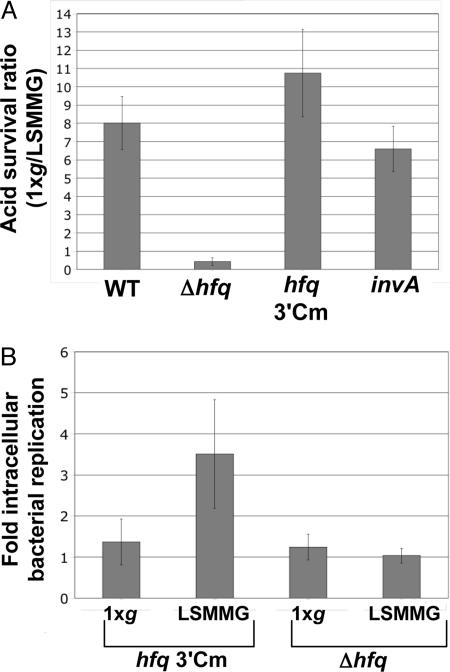
LSMMG-induced alterations in acid stress resistance and macrophage survival of S. typhimurium have previously been shown to be associated with global changes in gene expression and virulence (5, 7). We grew WT and isogenic hfq mutant strains of S. typhimurium in the RWV in the LSMMG and 1 × g positions and assayed the acid stress response and macrophage survival of these cultures. Whereas the WT strain displayed a significant difference in acid resistance between the LSMMG and 1 × g cultures, this response was not observed in the hfq mutant, which contains a deletion of the hfq gene and replacement with a Cm-r cassette (Fig. 2A). Two control strains, hfq 3′Cm (containing an insertion of the Cm-r cassette just downstream of the WT hfq gene) and invA kanamycin (Km) (containing a Km-r insertion in a gene unrelated to stress resistance), gave the same result as the WT strain. We also observed increased intracellular replication of the LSMMG-grown WT (hfq 3′Cm) strain in infected J774 macrophages as compared with the 1 × g control, and this phenotype was not observed in the hfq mutant strain (Fig. 2B). Collectively, these data indicate that Hfq is involved in the bacterial space flight response as confirmed in a ground-based model of microgravity conditions. In addition, the intracellular replication phenotype inside macrophages correlates with the finding that space flight and LSMMG cultures exhibit increased virulence in mice (see next paragraph and ref. 5).
Fig. 2.
Hfq is required for S. typhimurium LSMMG-induced phenotypes in RWV culture. (A) The survival ratio of WT and isogenic hfq, hfq 3′Cm, and invA mutant strains in acid stress after RWV culture in the LSMMG and 1 × g positions is plotted (P < 0.05, ANOVA). (B) Fold intracellular replication of S. typhimurium strains hfq 3′Cm and Δhfq in J774 macrophages after RWV culture as above. Intracellular bacteria were quantitated at 2 h and 24 h after infection, and the fold increase in bacterial numbers between those two time periods was calculated (P < 0.05, ANOVA).
Increased Virulence of S. typhimurium Grown in Space Flight as Compared with Ground Controls.
Because growth during space flight and potential global reprogramming of gene expression in response to this environment could alter the virulence of a pathogen, we compared the virulence of S. typhimurium space flight samples to identical ground controls as a second major part of our study. Bacteria from flight and ground cultures were harvested and immediately used to inoculate female BALB/c mice via a per-oral route of infection on the same day as the Shuttle landing. Two sets of mice were infected at increasing dosages of either flight or ground cultures, and the health of the mice was monitored every 6–12 h for 30 days. Mice infected with bacteria from the flight cultures displayed a decreased time to death (at the 107 dosage), increased percent mortality at each infection dosage, and a decreased LD50 value compared with those infected with ground controls (Fig. 1 B–D). These data indicate increased virulence for space flight S. typhimurium samples and are consistent with previous studies in which the same strain of S. typhimurium grown in the RWV under LSMMG conditions displayed enhanced virulence in a murine model as compared with 1 × g controls (5).
SEM of Space Flight and Ground Cultures.
To determine any morphological differences between flight and ground cultures, SEM analysis of bacteria from these samples was performed. Although no difference in the size and shape of individual cells in both cultures was apparent, the flight samples demonstrated clear differences in cellular aggregation and clumping that was associated with the formation of an extracellular matrix (Fig. 1E). Consistent with this finding, several genes associated with surface alterations related to biofilm formation changed expression in flight (up-regulation of wca/wza colonic acid synthesis operon, ompA, fimH; down-regulation of motility genes) (Table 1). SEM images of other bacterial biofilms show a similar matrix accumulation (20, 21). Because extracellular matrix/biofilm formation can help to increase survival of bacteria under various conditions, this phenotype indicates a change in bacterial community potentially related to the increased virulence of the flight bacteria in the murine model.
Discussion
To our knowledge, these results represent the first documented gene expression changes that occur in bacterial cells (and any microbial pathogen) during space flight and accordingly demonstrate that a microgravity growth condition provides an environmental signal that can induce molecular changes in bacterial cells. To our knowledge, these results also provide the first direct evidence that growth during space flight can alter the virulence of a pathogen; in this study, S. typhimurium grown in space flight displayed increased virulence in a murine infection model compared with identical ground controls. Importantly, these results correlate with previous findings in which the same strain of S. typhimurium displayed increased virulence in the murine model after growth in the low-shear microgravity-like conditions of the RWV bioreactor. In agreement with the increased virulence observed in the space flight samples, bacteria cultured in flight exhibited cellular aggregation and extracellular matrix formation consistent with biofilm production. Moreover, several Salmonella genes associated with biofilm formation changed expression in flight. In addition, the space flight analogue culture environment of the RWV was used to verify a mechanistic role for Hfq as a global regulator of microbial responses during growth in low-shear microgravity-like growth conditions similar to those found in space flight liquid culture.
Strategies designed to counteract the virulence-enhancing effects of space flight in microbes provide important potential benefits to crew health and open insight into novel antimicrobial strategies on Earth. Accordingly, the identification of global regulators, such as Hfq, that coordinate microbial responses to these biomedically relevant environments provides targets at which these strategies can be directed. Hfq is an RNA-binding global regulatory protein that is conserved in a wide range of organisms, both prokaryotes and eukaryotes, and primarily acts as a chaperone to stabilize interactions between small RNA and mRNA molecules (14). Further study is needed to determine whether changes in Hfq regulon expression under space flight and LSMMG space flight analogue conditions alter critical RNA interactions that control virulence and other microbial responses. Because low fluid shear is encountered by pathogens in the host, these responses may be important for bacterial reprogramming during transitions between environments of different physiological fluid shear levels (such as from the flow of a lumen to the protected environment between brush border microvilli), which results in enhanced survival and infection.
Materials and Methods
Strains, Media, and Chemical Reagents.
The virulent, mouse-passaged S. typhimurium derivative of SL1344 termed χ3339 was used as the WT strain in all flight- and ground-based experiments (5). Isogenic derivatives of SL1344 with mutations Δhfq, hfq 3′Cm, and invA Km were used in ground-based experiments (13, 22). The Δhfq strain contains a deletion of the hfq ORF and replacement with a chloramphenicol resistance cassette, and the hfq 3′Cm strain contains an insertion of the same cassette immediately downstream of the WT hfq ORF. The invA Km strain contains a Km resistance cassette inserted in the invA ORF. Lennox broth was used as the growth medium in all experiments (5), and PBS (Invitrogen, Carlsbad, CA) was used to resuspend bacteria for use as inoculum in the FPAs. The RNA fixative RNA Later II (Ambion, Austin, TX), glutaraldehyde (16%; Sigma, St. Louis, MO), and formaldehyde (2%; Ted Pella, Redding, CA) were used as fixatives in flight experiments.
Loading of FPA.
An FPA consists of a glass barrel that can be divided into compartments (by means of the insertion of rubber stoppers) and a lexan sheath into which the glass barrel is inserted (see SI Fig. 4). Each compartment in the glass barrel was filled with a solution in an order such that the solutions would be mixed at specific time points in flight via two actions: (i) downward plunging action on the rubber stoppers and (ii) passage of the fluid in a given compartment through a bevel on the side of the glass barrel such that it was released into the compartment below. Glass barrels and rubber stoppers were coated with a silicone lubricant (Sigmacote; Sigma) and autoclaved separately before assembly. A stopper with a gas-exchange membrane was inserted just below the bevel in the glass barrel before autoclaving. FPA assembly was performed aseptically in a laminar flow hood in the following order: 2.0 ml of Lennox broth medium on top of the gas-exchange stopper, one rubber stopper, 0.5 ml of PBS containing bacterial inoculum (≈6.7 × 106 bacteria), another rubber stopper, 2.5 ml of either RNA fixative or Lennox broth medium, and a final rubber stopper. Syringe needles (gauge 25 5/8) were inserted into rubber stoppers during this process to release air pressure and facilitate assembly. To facilitate group activation of FPAs during flight and to ensure proper containment levels, sets of eight FPAs were loaded into larger containers termed group activation packs.
Murine Infection Assay.
Six- to 8-week-old female BALB/c mice (housed in the Animal Facility at the Space Life Sciences Laboratory at Kennedy Space Center) were fasted for ≈6 h and then per-orally infected with increasing dosages of S. typhimurium harvested from flight and ground FPA cultures and resuspended in buffered saline gelatin (5). Ten mice per infectious dosage were used, and food and water were returned to the animals within 30 min after infection. The infected mice were monitored every 6–12 h for 30 days. The LD50 value was calculated by using the formula of Reed and Muench (23).
SEM.
A portion of cells from the viable, media-supplemented cultures from flight and ground FPAs was fixed for SEM analysis by using 8% glutaraldehyde and 1% formaldehyde and was processed for SEM as described previously (24).
Microarray Analysis.
Total cellular RNA purification, preparation of fluorescently labeled, single-stranded cDNA probes, probe hybridization to whole-genome S. typhimurium microarrays, and image acquisition were performed as previously described (7) using three biological and three technical replicates for each culture condition. Flow cytometric analysis revealed that cell numbers in flight and ground biological replicate cultures were not statistically different (using SYTO-BC dye per the manufacturer's recommendations; Invitrogen). Data from stored array images were obtained with QuantArray software (Packard Bioscience, Billerica, MA) and statistically analyzed for significant gene expression differences by using the Webarray suite as described previously (25). GeneSpring software was also used to validate the genes identified with the Webarray suite. To obtain the genes comprising the space flight stimulon as listed in SI Table 2, the following parameters were used in Webarray: a fold increase or decrease in expression of 2-fold or greater, a spot quality (A value) of >9.5, and P value of <0.05. For some genes listed in Table 1, the following parameters were used: a fold increase or decrease in expression of value >1.6 or <0.6, respectively, an A value of 8.5 or greater, and P value of <0.1. The vast majority of genes listed in Table 1 had an A value of >9.0 (with most being >9.5) and a P value of 0.05 or less. The exceptions were as follows: sbmA (P = 0.06), oxyS (A = 8.81), rplY (A = 8.95), cspD (A = 8.90), yfiA (P = 0.08), ompX (P = 0.09), hns (P = 0.08), rmf (A = 8.82), wcaD (P = 0.09), and fliE (A = 8.98). To identify space flight stimulon genes also contained in the Hfq regulon, proteins or genes found to be regulated by Hfq or RNAs found to be bound by Hfq as reported in the indicated references were scanned against the space flight microarray data for expression changes within the parameters above (8, 12, 13, 16, 26).
Multidimensional Protein Identification Analysis via Tandem MS Coupled to Dual Nano-Liquid Chromatography.
Acetone–protein precipitates from whole-cell lysates obtained from flight and ground cultures (representing three biological replicates) were subjected to multidimensional protein identification analysis using the tandem MS–dual nano-liquid chromatography technique as described previously (27, 28). Tandem MS spectra of peptides were analyzed with TurboSEQUEST version 3.1 and XTandem software, and the data were further analyzed and organized by using the Scaffold program (29, 30). Please see SI Table 3 for the specific parameters used in Scaffold to identify the proteins in this study.
Ground-Based RWV Cultures and Acid Stress and Macrophage Survival Assays.
S. typhimurium cultures were grown in the RWV in the LSMMG and 1 × g orientations and assayed for resistance to pH 3.5 and survival inside J774 macrophages as described previously (5), except that the RWV cultures were grown for 24 h at 37°C. For acid stress assays, the percentage of surviving bacteria present after 45–60 min of acid stress (compared with the original number of bacteria before addition of the stress) was calculated. A ratio of the percent survival values for the LSMMG and 1 × g cultures was obtained (indicating the fold difference in survival between these cultures) and is presented as the acid survival ratio in Fig. 2A. The mean and standard deviation from three independent experimental trials are presented. For macrophage survival assays, the number of bacteria present inside J774 macrophages at 2 h and 24 h after infection was determined, and the fold difference between these two numbers was calculated. The mean and standard deviation of values from three independent experimental trials (each done in triplicate tissue culture wells) are presented. The statistical differences observed in the graphs in Fig. 2 were calculated at P < 0.05.
Supplementary Material
Acknowledgments
We thank all supporting team members at Kennedy Space Center, Johnson Space Center, Ames Research Center, Marshall Space Flight Center, and BioServe Space Technologies; the Crew of STS-115; and Neal Pellis, Roy Curtiss III, Marc Porter, Brian Haight, Shawn Watts, Michael Caplan, Joseph Caspermeyer, Clint Coleman, and Charles Arntzen. This work was supported by National Aeronautics and Space Administration Grant NCC2-1362 (to C.A.N.), Louisiana Board of Regents Grant NNG05GH22H (to C.B.-C.), the Arizona Proteomics Consortium (supported by National Institute on Environmental Health Sciences Grant ES06694 to the Southwest Environmental Health Sciences Center), National Institutes of Health/National Cancer Institute Grant CA023074 (to the Arizona Cancer Center), and the BIO5 Institute of the University of Arizona.
Abbreviations
- LSMMG
low-shear modeled microgravity
- RWV
rotating wall vessel
- FPA
fluid processing apparatus
- Km
konamycin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE8573).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707155104/DC1.
References
- 1.Grigoriev AI, Svetaylo EN, Egorov AD. Environ Med. 1998;42:83–94. [PubMed] [Google Scholar]
- 2.Sonnenfeld G, Shearer WT. Nutrition. 2002;18:899–903. doi: 10.1016/s0899-9007(02)00903-6. [DOI] [PubMed] [Google Scholar]
- 3.Taylor GR. J Leukocyte Biol. 1993;54:202–208. doi: 10.1002/jlb.54.3.202. [DOI] [PubMed] [Google Scholar]
- 4.Taylor GR, Konstantinova I, Sonnenfeld G, Jennings R. Adv Space Biol Med. 1997;6:1–32. doi: 10.1016/s1569-2574(08)60076-3. [DOI] [PubMed] [Google Scholar]
- 5.Nickerson CA, Ott CM, Mister SJ, Morrow BJ, Burns-Keliher L, Pierson DL. Infect Immun. 2000;68:3147–3152. doi: 10.1128/iai.68.6.3147-3152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, Pierson DL. Microbiol Mol Biol Rev. 2004;68:345–361. doi: 10.1128/MMBR.68.2.345-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson JW, Ramamurthy R, Porwollik S, McClelland M, Hammond T, Allen P, Ott CM, Pierson DL, Nickerson CA. Proc Natl Acad Sci USA. 2002;99:13807–13812. doi: 10.1073/pnas.212387899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueroa-Bossi N, Lemire S, Maloriol D, Balbontin R, Casadesus J, Bossi L. Mol Microbiol. 2006;62:838–852. doi: 10.1111/j.1365-2958.2006.05413.x. [DOI] [PubMed] [Google Scholar]
- 9.Masse E, Gottesman S. Proc Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muffler A, Traulsen DD, Fischer D, Lange R, Hengge-Aronis R. J Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vecerek B, Moll I, Afonyushkin T, Kaberdin V, Blasi U. Mol Microbiol. 2003;50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 12.Guisbert E, Rhodius VA, Ahuja N, Witkin E, Gross CA. J Bacteriol. 2007;189:1963–1973. doi: 10.1128/JB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sittka A, Pfeiffer V, Tedin K, Vogel J. Mol Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valentin-Hansen P, Eriksen M, Udesen C. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 15.Valentin-Hansen P, Johansen J, Rasmussen AA. Curr Opin Microbiol. 2007;10:152–155. doi: 10.1016/j.mib.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Will WR, Frost LS. J Bacteriol. 2006;188:124–131. doi: 10.1128/JB.188.1.124-131.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond TG, Hammond JM. Am J Physiol. 2001;281:F12–F25. doi: 10.1152/ajprenal.2001.281.1.F12. [DOI] [PubMed] [Google Scholar]
- 18.Nickerson CA, Ott CM. ASM News. 2004;70:169–175. [Google Scholar]
- 19.Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, LeBlanc CL, Honer zu Bentrup K, Hammond T, Pierson DL. J Microbiol Methods. 2003;54:1–11. doi: 10.1016/s0167-7012(03)00018-6. [DOI] [PubMed] [Google Scholar]
- 20.Little B, Wagner P, Ray R, Pope R, Scheetz R. J Ind Microbiol. 1991;8:213–222. [Google Scholar]
- 21.Priester JH, Horst AM, Van de Werfhorst LC, Saleta JL, Mertes LA, Holden PA. J Microbiol Methods. 2007;68:577–587. doi: 10.1016/j.mimet.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 22.Wilson JW, Nickerson CA. BMC Evol Biol. 2006;6:2. doi: 10.1186/1471-2148-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed LJ, Muench H. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 24.Nickerson CA, Goodwin TJ, Terlonge J, Ott CM, Buchanan KL, Uicker WC, Emami K, LeBlanc CL, Ramamurthy R, Clarke MS, et al. Infect Immun. 2001;69:7106–7120. doi: 10.1128/IAI.69.11.7106-7120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 26.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 27.Cooper B, Eckert D, Andon NL, Yates JR, Haynes PA. J Am Soc Mass Spectrom. 2003;14:736–741. doi: 10.1016/S1044-0305(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 28.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, II, Smith RD. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 29.Craig R, Beavis RC. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 30.Eng JK, McCormack AL, Yates JR. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.