Abstract
A key factor in the development of Type II diabetes is the loss of insulin producing pancreatic β-cells. The amyloidogenic human Islet Amyloid Polypeptide (hIAPP also known as human amylin) is believed to play a crucial role in this biological process. Previous studies have shown that hIAPP forms small aggregates that kill β-cells by disrupting the cellular membrane. In this study, we report membrane fragmentation by hIAPP using solid-state NMR experiments on nanotube arrays of anodic aluminum oxide containing aligned phospholipid membranes. In a narrow concentration range of hIAPP, an isotropic 31P chemical shift signal indicative of the peptide-induced membrane fragmentation was detected. Solid-state NMR results suggest that membrane fragmentation is related to peptide aggregation as the presence of Congo Red, an inhibitor of amyloid formation, prevented membrane fragmentation and the non-amyloidogenic rat-IAPP did not cause membrane fragmentation. The disappearance of membrane fragmentation at higher concentrations of hIAPP suggests an alternate kinetic pathway to fibril formation in which membrane fragmentation is inhibited.
Increasing evidence links the toxicity of human Islet Amyloid Polypeptide (hIAPP) found in pancreatic β-cells to the development of Type II Diabetes.1 Soluble oligomers of hIAPP have been shown to kill β-cells by binding to and disrupting the cellular membrane.2-4 Interestingly, other amyloidogenic peptides such as Aβ, α-synuclein, and polyglutamine have also been found to cause cell death by a similar mechanism.5 Therefore, there is considerable interest in investigating the interaction of amyloidogenic peptides with phospholipid membranes.
Human-IAPP (also known as human amylin) has been shown to be toxic to β-cells while the rat version of IAPP (rIAPP) is known to be non-toxic.2-4 A comparison of sequences between mammals such as humans that develop type 2 diabetes and mammals such as rats that do not develop type 2 diabetes revealed important differences in a ten amino acid stretch in the segment 20-29 (SNNFGAILSS in the human sequence, SNNLGPVLPP in the rat sequence). This segment has also been shown to be critical for the aggregation of the peptides into amyloid fibers, studies have shown that this fragment of hIAPP (hIAPP20-29) is amyloidogenic whereas the 20-29 fragment of rIAPP (rIAPP20-29) is non-amyloidogenic.6-10 Furthermore, full-length hIAPP containing proline mutations in the 20-29 segment is rendered non-amyloidogenic, illustrating the critical role of this sequence for fibrilogenesis.8 Like full-length hIAPP, hIAPP20-29 inserts into lipid bilayers, albeit with a lower binding affinity and without the preference for anionic lipids shown by full length hIAPP.11
In this study, we report membrane fragmentation by the amyloidogenic hIAPP20-29 fragment and not by the corresponding rIAPP sequence using solid-state 31P NMR spectroscopy. The hIAPP20-29 and rIAPP20-29 peptides were synthesized by standard FMOC methods with an amidated C-terminus and an acetylated N-terminus. Purity was measured as greater than 95% by analytical HPLC and the sequence was verified by electrospray MAS. Membrane fragmentation was detected by the presence of a motionally averaged peak at the isotropic 31P chemical shift frequency. Bilayers of DMPC (1,2-Dimyristoyl-sn-Glycero-3-Phosphocholine) were used as model membranes to investigate the membrane interaction of hIAPP20-29 and rIAPP20-29. As previously utilized in studies on antimicrobial peptides12, aligned phospholipid bilayers were used to determine the mechanism of membrane disruption by IAPP. Since the preparation of mechanically aligned bilayers takes several days and the sample lacks bulk water, in this study, we chose to use anodic aluminum oxide (AAO) nanotubes to rapidly align bilayers with excess water. Previous studies have demonstrated that the alignment of lipid bilayers in AAO nanotubes is suitable for solid-state NMR experiments.13-15 As described elsewhere, aligned samples are useful to measure peptide-induced structural disorders in lipid bilayers using solid-state NMR experiments.16
To assess membrane fragmentation, AAO discs containing 4 mg of DMPC and between 0 to 4 mole % of either hIAPP20-29 or rIAPP20-29 were examined in increments of 0.25 mole % peptide by 31P NMR. Samples for 31P NMR were prepared by the deposition of multilamellar vesicles (MLVs) onto AAO discs. MLV samples were prepared by mixing a stock solution of the peptide in methanol (1mg/ml) with 200 μl of a choloroform solution of DMPC (20 mg/ml). The solvent was removed from the peptide/lipid samples by first drying the sample under a stream of nitrogen and then by drying under a vacuum overnight to completely remove any residual solvent. The resulting peptide/lipid film was directly hydrated with 70 μl of Hepes buffer (50 mM, pH 7.4) to form multilamellar vesicles. The MLV sample was heated above the transition temperature of DMPC and then pipetted onto a single 19 mm radius AAO disc cut into 10 mm by 10 mm strips to fit into a 14 × 14 × 4 mm flat-coil probe. The sample was then sealed in a plastic bag along with extra buffer (∼200 μl) to prevent dehydration during signal acquisition. The lipid bilayers are estimated to occupy approximately 14% of the pore volume in the AAO nanotube array (the remainder is filled with buffer).
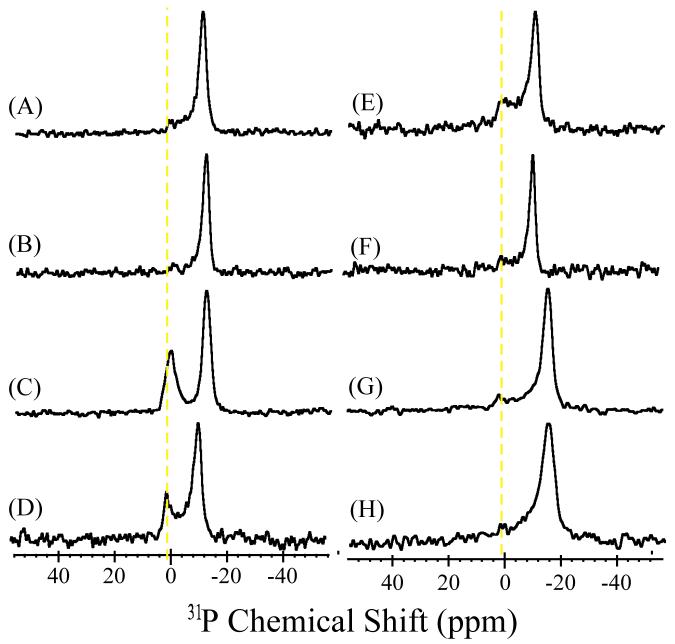
Representative 31P chemical shift spectra of DMPC bilayers containing varying concentrations of hIAPP20-29 and rIAPP20-29 are shown in Figure 1. To ensure reproducibility, spectra were acquired for 3 independent samples at each concentration. The spectra were acquired on a Chemagnetics/Varian 400 MHz spectrometer using a flat-coil (14 × 14 × 4 mm) probe and a spin-echo sequence (90°-τ-180°-τ, τ= 60 μs) with the bilayer normal perpendicular to the magnetic field at 37°C. 31P spectra were referenced relative to liquid H3PO4. For each sample 2048 transients were collected with a recycle delay of 3 seconds and 60 Hz line broadening. The nanotube material itself has a broad resonance (∼20 ppm wide) centered ∼10 ppm.15 This signal was subtracted for clarity.
Figure 1.
Phosphorus-31 chemical shift spectra of aligned DMPC bilayers containing various mole % of IAPP in AAO nanotubes: (A) 0%, (B) 0.5% hIAPP20-29, (C) 0.75 % hIAPP20-29, (D) 1% hIAPP20-29, (E) 1% hIAPP20-29 after incubation for 48 hours, (F) 1.25% hIAPP20-29, (G) 0.5% rIAPP20-29, (H) 1% rIAPP20-29.
A single peak at -13.0 ppm in the 31P chemical shift spectrum is observed for pure lipids indicating an unperturbed lamellar phase lipid bilayer with a high degree of alignment in the magnetic field (Figure 1A).13 A low intensity broad signal can be due to the unaligned lipids. A small shift of the main peak (<0.5 ppm for hIAPP20-29 (Figures 1B and 1C) and ∼3 ppm for rIAPP20-29 (Figures 1G and 1H)) was observed. This could suggest that some of the peptides bind to the lipid head group region, which could result in a change in the conformation of the lipid head group as has been reported for other membrane surface binding peptides.17 The 31P spectra suggest that at low (up to 0.75%) (Figure 1B) and high (>1.25%) (Figure 1F) mole % of hIAPP20-29 lipid bilayers were in the lamellar phase with no observable disorder induced by the peptide. However, within a narrow range of hIAPP20-29 mole % (∼0.75% to 1.25%, Figures 1C&D), an isotropic peak near -1 ppm was observed suggesting peptide-induced membrane fragmentation in lipid bilayers. A slight deviation of this peak frequency from the isotropic chemical shift and the line width (full width at half maximum ranges from ∼630 to ∼1090 Hz) could be due to restricted motion and/or residual alignment of bilayer fragments in nanopores. The isotropic peak was still observable even after incubation for 48 hours or one week (Figure 1E), but the lineshape showed some variations. It should be noted that Congo Red staining on large unilamellar vesicles incubated for 48 hours revealed the presence of amyloid formation (data not shown); similar results have been reported in the literature.6 On the other hand, as seen from the representative 31P spectra (Figures 1G&H), the non-amyloidogenic rIAPP20-29 peptide did not fragment lipid bilayers at any of the concentrations examined (0.25 to 3 mole %).
Further confirmation of the presence of small vesicle-like structures was obtained by dynamic light scattering measurements. MLV peptide-lipid samples (10 mg) were prepared as above then centrifuged at 24,000 g for 2 hours. The supernatant was separated from the lipid pellet and the vesicle size in the supernatant measured with a NICOMP 380 particle sizer. At 1% peptide, the particles were determined to be polydisperse with an average diameter of 103 nm and a polydispesity of 86 nm. The approximate size of the vesicle-like structures is in agreement with a previous report on the formation of similar non-bilayer structures with the Aβ1-42 peptide although the polydispersity is much greater.18 No appreciable light scattering was detected from the rIAPP20-29 samples or a control lipid sample without peptide.
The absence of the isotropic 31P peak at higher concentrations of hIAPP20-29 and the inability of non-amyloidogenic rIAPP20-29 to fragment the membrane indicate that peptide aggregation on the membrane plays a crucial role in this process. To show the correlation between the aggregation of hIAPP and membrane fragmentation, Congo Red was added to the hydration buffer of the sample. Congo Red is known to inhibit the formation of amyloid structure, most likely by interfering with the π-stacking interactions between peptides necessary for amyloidogenesis.19 In contrast to some other inhibitors such as rifampicin, Congo Red does not prevent the binding of IAPP to the membrane.20 As shown in Figure 2, Congo Red suppresses the isotropic peak formation, incompletely at 2 mole % (Figure 2B) and completely at 5 mole % (Figure 2C) of Congo Red. This suggests that the isotropic peak is related to the potential of the peptide to form aggregates.
Figure 2.
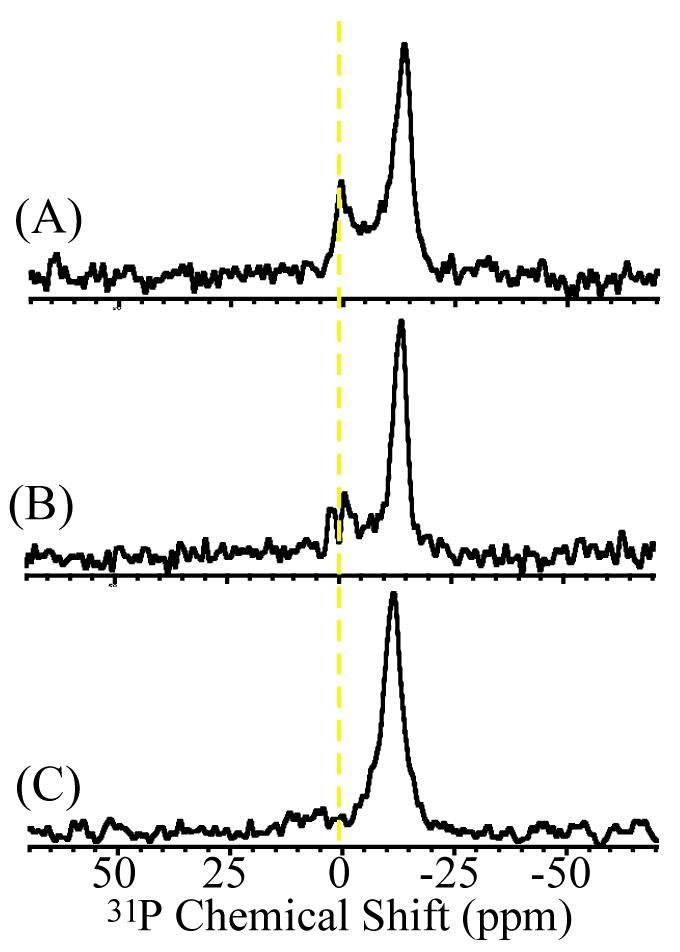
31P chemical shift spectra of DMPC bilayers containing 1 mole % of hIAPP aligned in AAO nanotubes with varying concentrations of Congo Red. (A) 0, (B) 2, and (C) 5 mole %. The isotropic peak, indicative of membrane fragmentation, is suppressed in the presence of Congo Red (B & C).
Remarkably, higher concentrations of hIAPP20-29 also suppress membrane fragmentation (Figures 1C & D). This is surprising because the standard detergent-like model for membrane fragmentation predicts membrane fragmentation after a threshold peptide/lipid ratio is reached and an increase in membrane fragmentation at higher peptide/lipid ratios.21 Since membrane permeabilization in hIAPP and other amyloid peptides is linked to the oligomeric state of the peptide, the lack of membrane fragmentation at higher peptide/lipid ratios suggests a different oligomeric state at higher peptide/lipid ratios. It is well known that the kinetics of fiber formation and the structures of amyloid fibers are dependant on the solution conditions of fiber growth. Numerous studies of amyloid proteins have shown the monomeric and fibrillar states of the protein are non-cytotoxic, however, transient oligomeric intermediates along the fibrilogenesis pathway show a strong cytotoxicity.2-4 If these cytotoxic intermediates along the fibrilogenesis pathway are responsible for the membrane fragmentation seen in Figures 1C and 1D, the concentration of intermediate states responsible for membrane fragmentation may be reduced either through an alteration of the initial distribution of hIAPP20-29 oligomeric states or a change in the kinetic pathway of fiber formation to disfavor the formation of cytotoxic oligomeric states at higher concentrations of hIAPP20-29. Further studies are in progress to characterize the lipid-peptide structures seen in Figures 1C and 1D. A schematic of possible pathways is illustrated in Figure 3.
Figure 3.
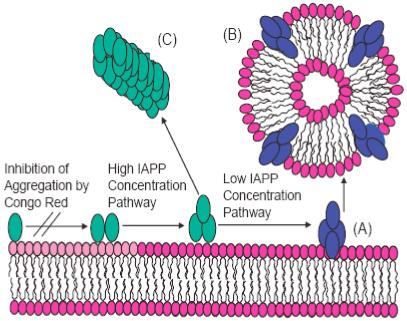
A schematic model for membrane fragmentation by hIAPP20-29. (A) At lower concentrations, the peptide aggregates on the lipid bilayer surface to form an intermediate state that is capable of extracting phospholipid molecules from the bilayer and leading to peptide-lipid vesicles (B). At higher concentrations, fiber formation follows an alternate pathway bypassing the formation of this intermediate (C).
A different mechanism of membrane disruption by amyloid proteins has been proposed in which lipid molecules are extracted directly from the membrane into the nascent amyloid fiber.22,23 If detached from the membrane, the amyloid fibers would tumble rapidly in solution and therefore the phospholipid molecules trapped within the fibers would give rise to a motionally averaged isotropic 31P signal. However, the high percentage of lipid in the isotropic phase at relatively low peptide/lipid ratios suggests this is not the mechanism primarily responsible for membrane fragmentation under these experimental conditions as it is unlikely the amyloid fiber can incorporate such a large excess of lipid molecules.
In agreement with our findings, membrane lesions have been detected in a previous study by atomic force microscopy upon the addition of the amyloidogenic human form of IAPP but not upon the addition of the non-amyloidogenic rat form of IAPP.24 However, the experimental setup used in that study could not distinguish between membrane lesions caused by a peptide-induced expansion of the bilayer and membrane lesions caused by micellization of the lipid bilayer. Furthermore, the membrane lesions caused by hIAPP spread from initial defects on the mica surface, leaving open the question of the role of surface defects on the mica surface in the membrane disruption process. In contrast, solid-state 31P NMR spectroscopy directly detects the loss of lamellar structure.
In summary, we have reported the direct detection of membrane fragmentation by the main amyloidogenic fragment of hIAPP using 31P solid-state NMR spectroscopy. One of the hallmarks of hIAPP cytotoxicity is the partial fragmentation of the cellular membrane into an abnormal mass of small vesicle-like structures.4 However, the existence of vesicle-like structures in electron micrographs of cells is by itself not conclusive evidence for direct fragmentation of the cell membrane by hIAPP. The presence of hIAPP in the cell may indirectly induce membrane fragmentation by activating other biochemical processes that lead to membrane fragmentation.25 However, the observation of membrane fragmentation in model phospholipid membranes by hIAPP20-29 reported in this study suggests that hIAPP may induce cytotoxicity by direct fragmentation of the cellular membrane.
Acknowledgment
This study was supported by research funds from NIH (AI054515 to A.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hoppener JW, Nieuwenhuis MG, Vroom TM, Lips CJ. Islet amyloid and diabetes mellitus type 2. New Eng. J. Med. 2000;144:1995–2000. [PubMed] [Google Scholar]
- 2.Konarkowska B, Aitken JF, Kistler J, Zhang SP, Cooper GJS. The aggregation potential of human amylin determines its cytotoxicity towards islet beta-cells. FEBS J. 2006;273:3614–3624. doi: 10.1111/j.1742-4658.2006.05367.x. [DOI] [PubMed] [Google Scholar]
- 3.Demuro A, Mina E, Kayed R, Milton SC, Parker I, Glabe CG. Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of souble amyloid oligomers. J. Biol. Chem. 2005;280:17294–17300. doi: 10.1074/jbc.M500997200. [DOI] [PubMed] [Google Scholar]
- 4.Janson J, Ashley RH, Harrison D, McIntyre S, Butler PC. The mechanism of Islet Amyloid Polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes. 1999;48:491–498. doi: 10.2337/diabetes.48.3.491. [DOI] [PubMed] [Google Scholar]
- 5.Lashuel HA, Lansbury PT. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Quart. Rev. Biophys. 2006;39:167–201. doi: 10.1017/S0033583506004422. [DOI] [PubMed] [Google Scholar]
- 6.Rhoades E, Agarwal J, Gafni A. Aggregation of an amyloidogenic fragment of human Islet Amyloid Polypeptide. Biochim. Biophys. Acta. 2000;1476:230–238. doi: 10.1016/s0167-4838(99)00248-4. [DOI] [PubMed] [Google Scholar]
- 7.Rhoades E, Gafni A. Micelle formation by a fragment of human Islet Amyloid Polypeptide. Biophys. J. 2003;84:3480–3487. doi: 10.1016/S0006-3495(03)70068-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westermark P, Engstrom U, Johnson KH, Westermark GT, Betsholtz C. Islet Amyloid Polypeptide: pinpointing amino acid residues linked to amyloid fibril formation. Proc. Natl. Acad. Sci. USA. 1990;87:5036–5040. doi: 10.1073/pnas.87.13.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mascioni A, Porcelli F, Ilangovan U, Ramamoorthy A, Venglia G. Conformational preferences of the amylin nucleation site in SDS micelles: An NMR study. Biopolymers. 2003;69:29–41. doi: 10.1002/bip.10305. [DOI] [PubMed] [Google Scholar]
- 10.Ilangovan U, Ramamoorthy A. Conformational studies of human Islet Amyloid Peptide using molecular dynamics and simulated annealing methods. Biopolymers. 1998;45:9–20. doi: 10.1002/(SICI)1097-0282(199801)45:1<9::AID-BIP2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 11.Engel MFM, Yagittop HA, Elgersma RC, Rijkers DTS, Liskamp RMJ, de Kruijff B, Hoppener JWM, Killian JA. Islet amyloid polypeptide inserts into phospholipid monolayers as monomer. J. Mol. Biol. 2006;356:783–78. doi: 10.1016/j.jmb.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 12.Ramamoorthy A, Thennarasu S, Lee DK, Tan A, Maloy L. Solid-State NMR investigation of the membrane-disrupting mechanism of antimicrobial peptides MSI-78 and MSI-594 derived from magainin 2 and melittin. Biophys. J. 2006;91:206–216. doi: 10.1529/biophysj.105.073890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorigan GA, Dave PC, Tiburu EK, Damodaran K, Abu-Baker S, Karp ES, Gibbons WJ, Minto RE. Solid-state NMR spectroscopic studies of an integral membrane protein inserted into aligned phospholipid bilayer nanotube arrays. J. Am. Chem. Soc. 2004;126:9504–9505. doi: 10.1021/ja047317+. [DOI] [PubMed] [Google Scholar]
- 14.Chekmenev EY, Hu J, Gorkov PL, Brey WW, Cross TA, Ruuge A, Smirnov AI. 15N and 31P solid-state NMR study of transmembrane domain alignment of M2 protein of influenza A virus in hydrated cylindrical lipid bilayers confined to anodic aluminum oxide nanopores. J. Mag. Reson. 2005;173:322–327. doi: 10.1016/j.jmr.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Gaede HC, Luckett KM, Polozov IV, Gawrisch K. Multinuclear NMR studies of single lipid bilayers supported in cylindrical aluminum oxide nanopores. Langmuir. 2004;20:7711–7719. doi: 10.1021/la0493114. [DOI] [PubMed] [Google Scholar]
- 16.Hallock KJ, Henzler Wildman KA, Lee DK, Ramamoorthy A. Sublimable solids can be used to mechanically align lipid bilayers for solid-state NMR studies. Biophys. J. 2002;82:2499. doi: 10.1016/S0006-3495(02)75592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramamoorthy A, Thennarasu S, Tan A, Lee DK, Clayberger C, Krensky AM. Cell selectivity correlates with membrane interactions: a case study on the antimicrobial peptide G15 derived from granulysin. BBA-Biomembranes. 2006;1758:154–163. doi: 10.1016/j.bbamem.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Lau TL, Ambroggio EE, Tew DJ, Cappai R, Masters CL, Fielio GD, Barnham KJ, Separovic F. Amyloid-β peptide disruption of lipid membranes and the effect of metal ions. J. Mol. Biol. 2006;356:759–770. doi: 10.1016/j.jmb.2005.11.091. [DOI] [PubMed] [Google Scholar]
- 19.Porat Y, Mazor Y, Erfat S, Gazit E. Inhibition of Islet Amyloid Polypeptide fibril formation: A potential role for heteroaromatic interactions. Biochemistry. 2004;43:14454–14462. doi: 10.1021/bi048582a. [DOI] [PubMed] [Google Scholar]
- 20.Harroun TA, Bradshaw JP, Ashley RH. Inhibitors can arrest the membrane activity of human Islet Amyloid Polypeptide independently of amyloid formation. FEBS Lett. 2001;507:200–204. doi: 10.1016/s0014-5793(01)02972-6. [DOI] [PubMed] [Google Scholar]
- 21.Bechinger B, Lohner K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta. 2006;1758:1529–1539. doi: 10.1016/j.bbamem.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Sparr E, Engel MF, Sakharov DV, Sprong M, Jacobs J, De Kruijff B, Hoppener JW, Killian JA. Islet Amyloid Polypeptide-induced membrane leakage involves uptake of lipids by forming amyloid fibers. FEBS Lett. 2004;577:117–120. doi: 10.1016/j.febslet.2004.09.075. [DOI] [PubMed] [Google Scholar]
- 23.Michikawa M, Gong JS, Fan QW, Sawamura N, Yanagisawa K. A novel action of Alzheimer’s amyloid β-protein (Aβ): Oligomeric Aβ promotes lipid release. J. Neurosci. 2001;21:7226–7235. doi: 10.1523/JNEUROSCI.21-18-07226.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green JD, Kreplak L, Goldsbury C, Blatter XL, Stolz M, Cooper GS, Seelig A, Kist-Ler J, Aebi U. Atomic force microscopy reveals defects within mica supported lipid bilayers induced by the amyloidogenic human amylin peptide. J. Mol. Biol. 2004;342:877–887. doi: 10.1016/j.jmb.2004.07.052. [DOI] [PubMed] [Google Scholar]
- 25.Trump BF, Croslev BP, Mergner GW. In: Cell Membranes: Biological and Pathological Aspects. Richter GW, Scarpelli DG, editors. Williams and Wilkins; Baltimore: 1971. pp. 84–126. [Google Scholar]