Abstract
A gradual loss of telomeric repeat sequences with aging previously has been noted in normal adult tissues, and this process has been implicated in cell senescence. No data exist that address the rate of telomere shortening in normal human cells within families or early in life. To address these questions, we measured telomere lengths in peripheral blood leukocytes (PBLs) from 75 members of 12 families and in a group of unrelated healthy children who were 5–48 months old. Here we report the surprising observation that rates of telomere attrition vary markedly at different ages. Telomeric repeats are lost rapidly (at a rate of >1 kilobase per year) from the PBLs of young children, followed by an apparent plateau between age 4 and young adulthood, and by gradual attrition later in life. These data suggest that the loss of telomeric repeats in hematopoietic cells is a dynamic process that is differentially regulated in young children and adults. Our results have implications for current models of how telomeric sequences are lost in normal somatic cells and suggest that PBLs are an excellent tissue to investigate how this process is controlled.
Eukaryotic chromosomes end in specialized nucleoprotein structures called telomeres whose integrity is maintained, at least in part, by the ribonucleoprotein enzyme telomerase (1, 2). It has been reported that telomeres in human germ cells and fetal tissues are more elongated than in adult tissues, which gradually lose telomeric DNA with aging (3–5). Telomerase activity is absent in most normal somatic cells, but is detected in normal proliferating cells and in the majority of cancers (reviewed in ref. 6). Taken together, these data and other recent experimental evidence implicate telomere attrition and telomerase as potentially important in the replicative senescence of normal somatic cells, and in the initiation and/or maintenance of the immortal phenotype that is one hallmark of neoplastic cells.
Recent observations have provided new insights into both the regulation of telomere length in normal cells and the phenotypic consequences of perturbing these processes. Certain mutations in yeasts that alter either the telomerase RNA or the DNA-sequence specific duplex binding protein RAP1 lead to massive telomere elongation that is directly proportional to loss of the ability of RAP1 to bind telomeric repeats or to form a higher order complex with them (7–9). The identification of a protein factor called TRF that specifically associates with the duplex DNA of human telomeres provided additional evidence that in human somatic cells such telomeric proteins, as well as telomerase, play an essential role in regulating the lengths of telomeric repeats (10). Blasco et al. (2) recently described the consequences of disrupting the murine gene encoding the RNA component of telomerase. Knockout mice were viable and fertile for the first few generations despite the complete absence of telomerase activity. There was a progressive loss of telomeric DNA from generation to generation and this was associated with an elevated frequency of chromosomal abnormalities after the third generation. However, cells from telomerase null mice gave rise to immortal cell lines in culture, could be transformed by oncogenes, and produced tumors in nude mice (2). Most recently, Bodnar and coworkers (11) found that ectopic expression of the catalytic subunit of telomerase in primary epithelial cells and fibroblasts markedly extended their lifespan in culture and repressed the expression of a marker of senescence. Taken together, such studies support a model whereby telomerase interacts with other cellular proteins to determine the length of telomeric repeats, and this, in turn, influences cell fate through an unknown signaling mechanism. However, the questions of whether normal human somatic cells actively regulate telomere length in vivo, or whether loss of repeat sequences occurs at a constant rate throughout life that is directly proportional to the number of cell divisions, have not been addressed directly.
Hematopoiesis involves the regulated production of differentiated blood cells of multiple lineages. The fates of immature blood cells are controlled by a complex set of interacting factors, including cell–cell interactions in the bone marrow microenvironment and the binding of soluble peptide growth factors to high-affinity receptors that are expressed at specific stages of hematopoietic development. Unlike many other somatic tissues, hematopoietic precursors proliferate in the bone marrow throughout life to maintain circulating numbers of white blood cells, erythocytes, and platelets. In addition, transplantation studies have established unequivocally that the bone marrows of normal adults contain multipotent precursors that are capable of fully reconstituting the hematopoietic system of recipients after myeloablative conditioning. For these reasons, and because blood cells can be obtained by a simple venipuncture procedure, the hematopoietic system previously has been used to examine telomere shortening and telomerase activity in normal tissues. Normal peripheral blood and bone marrow cells express low levels of telomerase activity (12–14). In one study, the highest telomerase activity was detected in immature bone marrow hematopoietic progenitors and telomerase was down-regulated with cellular maturation (14). Leukemic blasts frequently show high levels of telomerase activity (12, 15) that are inversely correlated with the length of telomeric repeats (12). However, this is not a consistent finding, and the telomerase activities of leukemic and normal hematopoietic cells show considerable overlap (12, 13, 15). Telomerase activity also is present in normal reactive lymph nodes and tonsils and in T lymphocytes that have been stimulated to divide by exposure to mitogens (16, 17). These findings are consistent with recent observations that telomerase activity correlates with proliferative status in normal nonhematopoietic human cells (18). Taken together, these data suggest that subsets of normal immature and proliferating hematopoietic cells of normal adults retain the capacity to produce telomerase.
The studies of telomere attrition in normal tissues reported to date primarily have focused on the decline that begins in early adult life and progresses gradually with advancing age (3–5). No previous work has examined whether the rate of telomere loss varies between families. In addition, although considerable loss of telomeric sequences is seen between fetal life and young adulthood; the kinetics of this decline are unknown. Here we show that rates of telomere attrition in blood cells are highly variable at different ages and are surprisingly rapid in young children. These data suggest that the process of telomere loss is differentially regulated in leukocytes from young children and adults and have implications for models of telomere dynamics in normal human somatic tissues.
MATERIALS AND METHODS
Patient Samples.
Umbilical cord blood samples were collected from the placental end of the cord after delivery. Specimens from adult participants were obtained by venipuncture. For the family studies, blood samples were available from the umbilical cord, the parents of the baby, and from at least two grandparents, great-grandparents or great-great-grandparents from the same kindred. In the studies to ascertain telomere attrition during the first few years of life, blood was obtained from healthy children who were undergoing phlebotomy for other reasons. The families were counseled regarding their participation, and informed consent was obtained before obtaining all blood samples. The study procedures were approved by the institutional review board at the Naval Hospital, Oakland, CA.
DNA Extraction and Southern Blotting.
After osmotic lysis to remove contaminating red blood cells, DNA was extracted from leukocyte pellets by standard methodology as previously described (19, 20). DNA samples prepared according to this protocol are from a mixed population of peripheral blood leukocytes (PBLs) that includes lymphocytes, monocytes, neutrophils, and eosinophils. The integrity of each DNA sample was confirmed by electrophoresis and visualization of an undigested 1-μg aliquot on an ethidium bromide-stained 0.8% agarose minigel. For analysis of telomeric and subtelomeric restriction fragment lengths, 10-μg aliquots of DNA were digested overnight with either BglII or StyI restriction endonuclease according to the manufacturer’s instructions (New England Biolabs). Electrophoresis was performed in 0.8% agarose gels at 80 V for 3–4 hr in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA) with constant recirculation of the buffer. A control lane of phage lambda DNA digested with HindIII was included in each gel, and the 2-kilobase (kb) lambda fragment was run out 12.5–15 cm from the origin. After electrophoresis, the gels were soaked in ethidium bromide and photographed, and the DNA was transferred to nylon membranes (Hybond, Amersham) for Southern blotting.
Probes.
Plasmids containing a 600-bp TTAGGG repeat insert and the TH2Δ subtelomeric fragment (21) were a generous gift of Titia DeLange (Rockefeller University, New York). These inserts were excised, gel-purified, and labeled to high specific activity with 32P-deoxy nucleotides by using random primers (for the TH2Δ fragment) or an AATCCC oligonucleotide (for the TTAGGG insert). After hybridization and washing, filters were subjected to autoradiography. Mean telomere and subtelomere lengths were calculated by directly measuring migration from the origin on the autoradiographs.
RESULTS
Telomere Lengths in Related Individuals from Multiple Generations.
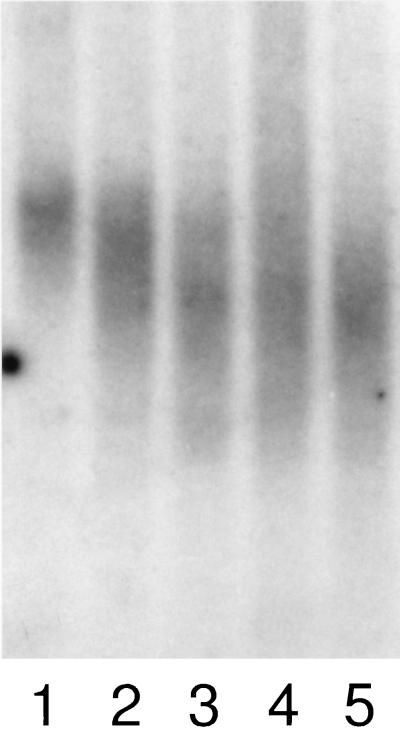
We first ascertained rates of telomeric shortening in PBLs from family members of different ages. We took advantage of the fact that newborn grandchildren often attract their grandparents to the hospital. This allowed us to compare telomere lengths in neonatal PBLs (isolated from the placental side of the umbilical cord) with paired blood samples from their healthy parents and grandparents. Table 1 presents the mean telomere lengths by generation for the 75 individuals from 12 kindreds who participated in the study. Data from one representative family are shown in Fig. 1. Mean telomere length declined from a mean ± 1 SD of 16.4 ± 1.2 kbs in the newborns, to 11.6 ± 1.2 kbs in their parents, to 9.6 ± 0.8 kbs in the grandparents (P < 0.001 for newborns vs. parents and P < 0.01 for parents vs. grandparents). Hence, although the average age difference between each generation was approximately 25 years (Table 1), the overall shortening of PBL telomere repeats was significantly greater between the newborns and their parents than between parents and grandparents (4.8 vs. 2.0 kbs). This pattern was consistent within families, and we did not detect significant variations between families (i.e., we did not identify families in whom the telomeric repeats were especially short or long). One interesting finding was that the telomeric repeats of neonatal leukocyte samples showed considerably less variability compared with the telomeric repeats of adult PBLs (Fig. 1).
Table 1.
Mean leukocyte telomere length in 12 multigenerational families
| Generation | Number of individuals | Mean age* and (range) | Mean telomere length (±1 SD) |
|---|---|---|---|
| Newborns | 12 | <1 h | 16.4 ± 1.2 kb |
| (0–1 h) | |||
| Parents | 24 | 26.5 years | 11.6 ± 1.2 kb |
| (20–36 years) | |||
| Grandparents | 35 | 50.4 years | 9.6 ± 0.8 kb |
| (42–72 years) | |||
| Great-grandparents† | 4 | 72 years | 8.0 ± 1.1 kb |
| (62–82 years) |
Mean age was calculated for the 68 individuals for whom this was recorded.
This group includes one great-great grandparent (age 82).
Figure 1.
Telomere repeats in a multigenerational family. Leukocyte DNA was digested with BglII and hybridized with a probe that recognizes the TTAGGG telomeric repeat. Lanes 1–5 contain DNA from umbilical cord of a newborn infant (1), his mother (2), father (3), maternal grandmother (4), and maternal grandfather (5). The wider vertical length in the bands seen in lanes 2–5 reflect greater variability in the length in individual telomeres in the leukocyte populations of adults relative to neonatal PBLs (lane 1).
Rapid Loss of Telomeric Repeat Sequences from the PBLs of Young Children.
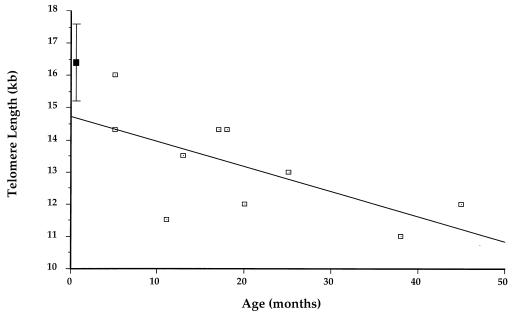
Because the most rapid decline in telomeric length was observed between newborns and parents, we wished to measure the rate of telomere shortening in PBLs during the first few years of life. The ideal experimental approach to this question involves obtaining serial blood samples by venipuncture from the same children; however, this approach was not acceptable to our Institutional Review Board because those individuals cannot give informed consent and the procedure offers no potential medical benefit to study participants. We therefore studied leukocyte DNA from a group of healthy young children who were having blood sampled for a medical indication to better define the slope of telomere shortening early in life. For these experiments, we obtained an aliquot of peripheral blood from 10 healthy children ≤ 48 months old who were having blood sampled for another reason. The mean telomere length was 13.2 ± 1.6 kbs. However, in this group a considerable decline in telomere length occurred over time. As shown in Fig. 2, the longest telomeric repeats were seen in children who were <18 months old, whereas the PBLs of older children had telomere lengths that were similar to those of adults. To ensure that telomere shortening resulted from selective loss of the terminal TTAGGG repeats, control experiments were performed in which patient DNA specimens were digested with StyI and hybridized with probe TH2Δ that recognizes a subtelomeric repeat sequence in a subset of human chromosomes (21). These experiments revealed a consistent subtelomeric fragment of approximately 4 kbs in individuals of all ages (data not shown). Hence, the observed shortening is attributable to terminal attrition of TTAGGG repeats rather than loss of subtelomeric sequences.
Figure 2.
Telomere lengths in 10 unrelated children between ages 5 and 24 months. ▪ shows the mean ± 1 SD for neonatal specimens. The R value of the regression line is 0.65, and the coefficient of variance (R2) is 0.424.
DISCUSSION
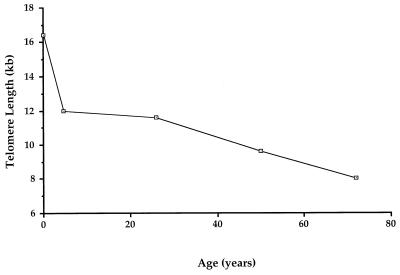
A major unresolved question in mammalian telomere biology involves whether loss of sequences from the ends of chromosomes in somatic cells occurs at a constant rate with each mitosis, or whether the extent of telomere loss varies over time because of changes in the cellular levels of positive and negative regulatory factors. The data from this study of normal PBLs support the latter model. We have made the surprising observation that loss of telomere sequences does not occur at a consistent rate in these cells throughout life. Instead, the data distinguish at least two phases of telomere shortening (Fig. 3). The first of these encompasses birth through about age 4 and is characterized by a rapid decline in average telomere length. Telomere lengths appear to remain stable until early adulthood (apparent stable phase). Like the initial phase of rapid shortening, this apparent plateau has not been recognized previously. It is seen during a time of extensive somatic growth and physical maturation. The final phase involves the previously reported gradual decline in mean telomere length that is associated with advancing age.
Figure 3.
Phases of telomere shortening in normal PBLs. The point shown for age 5 is taken from the oldest child shown in Fig. 2. The initial phase is characterized by rapid loss of telomeric repeats. An apparent stabilization then occurs between age 5 and young adulthood. Telomere loss resumes at a slower rate later as adults grow older. See text for further discussion.
The stabilization in telomere length seen between the middle of the first and third decades cannot be explained by postulating that most leukocyte cell divisions already have occurred by age 4. Circulating PBLs are lost throughout life and are constantly replaced by new cells generated from the proliferation and differentiation of hematopoietic progenitors. Because the total number of circulating blood cells correlates with body mass, an average adult has 3–5 times as many PBLs as a 4-year-old. Similarly, an average 4-year-old has 4–5 times as many PBLs as a newborn. In addition, preliminary data from patients with solid tumors treated with the hematopoietic growth factor granulocyte-macrophage colony stimulating factor, which rapidly and dramatically increases circulating leukocyte counts caused by increased production by the bone marrow, revealed no changes in telomere lengths (R.W.F. and K.M.S., unpublished observations).
We considered the possibility that the rapid loss of telomeric sequences in PBLs we observed early in life reflects qualitative differences in the cellular composition of hematopoietic tissues between young children and adults. Umbilical cord and neonatal blood are enriched for immature hematopoieitic progenitors relative to adult blood (22); this has led to the development of cord blood banks for bone marrow transplantation (23). However, this explanation appears unlikely. First, although it is possible that these immature subpopulations have very long telomeric repeats, their numbers in the circulation are insufficient to explain the large drop in mean telomere lengths that we found. In addition, we observed relatively less variation in the lengths of telomeric repeats in neonatal samples; this finding does not support a major contribution from a subset of immature cells with very long telomeric repeats. It is possible that the rapid loss of telomeric DNA early in life is caused by a high rate of proliferation in the most immature subsets of hematopoietic progenitors. According to this model, the large numbers of cell divisions that are required to sustain hematopoiesis throughout life would be initiated from a partially differentiated pool of progenitors that already have undergone a substantial amount of telomeric shortening. Our data are also consistent with a model whereby telomere sequences are not lost at the same rate in each cell division, but that this process is regulated by changing levels of a competing set of positive and negative factors throughout life (24). A provocative finding of Bodnar et al. (11) is that ectopic overexpression of the catalytic subunit of telomerase often was associated with an increase in the length of telomeric repeats over time in proliferating cultures of primary cells. This raises the possibility that some normal somatic cells are capable of reversing the normal decline in telomeric length under certain circumstances. Although there are no data that speak directly to this question, immature hematopoietic cells are known to express telomerase and there is massive proliferation in the repopulating bone marrow after transplantation. This clinical setting might provide an opportunity to test the hypothesis that a normal somatic cell population can up-regulate telomerase and undergo a net gain in telomeric sequences in certain in vivo situations. Regardless of the mechanism, our data demonstrate developmental stage-dependent changes in rates of telomere loss and underscore the complex nature of this process in normal human somatic cells.
Acknowledgments
We are indebted to Dr. Titia de Lange for providing us with plasmids containing the TTAGGG repeat and TH2Δ probes. This work was supported, in part, by the U.S. Navy Clinical Investigation Program, by a grant from the Frank A. Campini Foundation, and by National Institutes of Health Grant GM26259.
ABBREVIATIONS
- PBL
peripheral blood leukocyte
- kb
kilobase
References
- 1.Blackburn E H. Nature (London) 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Blasco M A, Lee H-W, Hande M P, Samper E, Lansdorp P M, DePinho R A, Greider C W. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 3.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 4.Hastie N D, Dempster M, Dunlop M G, Thompson A M, Green D K, Allshire R C. Nature (London) 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 5.Lindsey J, McGill N I, Lindsey L A, Green D K, Cooke H J. Mutat Res. 1991;256:45–48. doi: 10.1016/0921-8734(91)90032-7. [DOI] [PubMed] [Google Scholar]
- 6.Greider C. Proc Natl Acad Sci USA. 1998;95:90–92. doi: 10.1073/pnas.95.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyrion G, Boakye K A, Lustig A J. Mol Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McEachern M J, Blackburn E H. Nature (London) 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 9.Krauskopf A, Blackburn E H. Nature (London) 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- 10.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. Science. 1995;270:1663–1666. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 11.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 12.Counter C M, Gupta J, Harley C B, Leber B, Bacchetti S. Blood. 1995;85:2315–2320. [PubMed] [Google Scholar]
- 13.Broccoli D, Young J W, de Lange T. Cell Biol. 1995;92:9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu C-P, Dragowska W, Kim N W, Vaziri H, Yui J, Thomas T E, Harley C B, Lansdorp P M. Stem Cells. 1996;14:239–248. doi: 10.1002/stem.140239. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Piatyszek M A, Kobayashi T, Estey E, Andreeff M, Deisseroth A B, Wright W E, Shay J W. Clin Cancer Res. 1996;2:799–803. [PubMed] [Google Scholar]
- 16.Brousset P, Al Saati T, Chaouche N, Zenou R-C, Schlaifer D, Chittal S, Delsol G. Blood. 1997;89:26–31. [PubMed] [Google Scholar]
- 17.Buchkovich K J, Greider C W. Mol Biol Cell. 1996;7:1443–1454. doi: 10.1091/mbc.7.9.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belair C D, Yeager T R, Lopez P M, Reznikoff C A. Proc Natl Acad Sci USA. 1997;94:13677–13682. doi: 10.1073/pnas.94.25.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon K M, Turhan A G, Chang S S Y, Bowcock A M, Rogers P C J, Carroll W L, Cowan M J, Glader B E, Eaves C J, Eaves A C, Kan Y W. J Clin Invest. 1989;84:984–989. doi: 10.1172/JCI114262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon K M, Turhan A G, Rogers P C J, Kan Y W. Genomics. 1992;14:121–125. doi: 10.1016/s0888-7543(05)80293-9. [DOI] [PubMed] [Google Scholar]
- 21.de Lange T, Shiue L, Myers R M, Cos D R, Naylor S L, Killery A M, Varmus H E. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broxmeyer H E, Douglas G W, Hangoc G. Proc Natl Acad Sci USA. 1989;86:3828–3832. doi: 10.1073/pnas.86.10.3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gluckman E, Rocha V, Boyer-Chammard A, Locatelli F, Arcese W, Pasquini R, Ortega J, Souillet G, Ferreira E, Laporte J-P, Fernandez M, Chastang C. N Engl J Med. 1997;337:373–381. doi: 10.1056/NEJM199708073370602. [DOI] [PubMed] [Google Scholar]
- 24.Lundblad V, Wright W E. Cell. 1996;87:369–375. doi: 10.1016/s0092-8674(00)81358-6. [DOI] [PubMed] [Google Scholar]