Abstract
Background
Although early-stage non-small-cell lung cancer (NSCLC) is considered a potentially curable disease following complete resection, patients have a wide spectrum of survival according to stage (IB, II, IIIA). Within each stage, gene expression profiles can identify patients with a higher risk of recurrence. We hypothesized that altered mRNA expression in nine genes could help to predict disease outcome: excision repair cross-complementing 1 (ERCC1), myeloid zinc finger 1 (MZF1) and Twist1 (which regulate N-cadherin expression), ribonucleotide reductase subunit M1 (RRM1), thioredoxin-1 (TRX1), tyrosyl-DNA phosphodiesterase (Tdp1), nuclear factor of activated T cells (NFAT), BRCA1, and the human homolog of yeast budding uninhibited by benzimidazole (BubR1).
Methodology and Principal Findings
We performed real-time quantitative polymerase chain reaction (RT-QPCR) in frozen lung cancer tissue specimens from 126 chemonaive NSCLC patients who had undergone surgical resection and evaluated the association between gene expression levels and survival. For validation, we used paraffin-embedded specimens from 58 other NSCLC patients. A strong inter-gene correlation was observed between expression levels of all genes except NFAT. A Cox proportional hazards model indicated that along with disease stage, BRCA1 mRNA expression significantly correlated with overall survival (hazard ratio [HR], 1.98 [95% confidence interval (CI), 1.11-6]; P = 0.02). In the independent cohort of 58 patients, BRCA1 mRNA expression also significantly correlated with survival (HR, 2.4 [95%CI, 1.01-5.92]; P = 0.04).
Conclusions
Overexpression of BRCA1 mRNA was strongly associated with poor survival in NSCLC patients, and the validation of this finding in an independent data set further strengthened this association. Since BRCA1 mRNA expression has previously been linked to differential sensitivity to cisplatin and antimicrotubule drugs, BRCA1 mRNA expression may provide additional information for customizing adjuvant antimicrotubule-based chemotherapy, especially in stage IB, where the role of adjuvant chemotherapy has not been clearly demonstrated.
Introduction
In 2006 in Europe, there were an estimated 386,300 lung cancer cases, with a substantially higher incidence in men than in women[1]. Among completely resected non-small-cell lung cancer (NSCLC) patients, 40% of stage I, 66% of stage II and 75% of stage IIIA patients die within five years of resection[2], and the benefit of adjuvant chemotherapy has not been demonstrated in stage IB. In the ANITA randomized trial, 5-year survival for patients with stage IB disease was 62% in the chemotherapy group and 64% in the control group; corresponding rates were 52% and 39% for stage II patients and 42% and 26% for stage IIIA[3]. In addition to disease stage, several studies have examined gene expression profiles in NSCLC, identifying molecular subtypes associated with patient outcome[4], [5], [6], [7]. Gene expression signatures ranging from five to 64 genes have been identified[6], [7], and cross-study comparisons have revealed significant, though incomplete, agreement of patterns predicting outcome[4]. Moreover, the ability to interpret the meaning of the individual genes in these signatures remains a challenge[8]. The gene expression signatures identified genes mostly related to cancer metastasis[6] but did not describe genes involved in DNA repair pathways.
Preclinical studies have demonstrated that a deficiency in any one of the more than 30 genes involved in the nucleotide excision repair (NER) pathway confers marked hypersensitivity to cisplatin[9]. Hypothesizing that elevated levels of NER genes could be not only predictive but also prognostic markers, we chose to examine the following genes, based on previous reports of their predictive value: excision repair cross-complementing 1 (ERCC1)[10], BRCA1[11], human homolog of yeast budding uninhibited by benzimidazole (BubR1)[12], [13], [14], myeloid zinc finger 1 (MZF1)[15], [16], [17], ribonucleotide reductase subunit M1 (RRM1)[18], [19], [20], thioredoxin-1 (TRX1)[21], [22], tyrosyl-DNA phosphodiesterase (Tdp1)[23], [24], [25]. In addition, we examined Twist[26], [27] and nuclear factor of activated T cells (NFAT)[28], [29], which are involved in the invasion-metastasis process. (Further details on the nine genes examined can be found in Text S1.)
None of these nine genes have been identified in gene expression profiles associated with NSCLC patient outcome[4], [5], [6], [7], with the exception of TRX1, which was identified by proteomic analysis and associated with poor survival[21]. In order to shed light on the prognostic value of these genes, we have examined their expression by real-time quantitative reverse transcriptase PCR (RT-PCR) in 126 completely resected NSCLC patients who did not receive adjuvant chemotherapy and correlated the results with survival.
Methods
Patients
NSCLC samples were obtained from 126 consecutive patients who underwent curative pulmonary resection at the Medical University of Gdansk (Gdansk, Poland) between 2000 and 2004, after obtaining approval from the institutional review board of the Medical University of Gdansk and patients' signed informed consent. The patients were 98 males and 28 females, with age at diagnosis ranging from 37 to 77 years (median age, 64 years). Seventy-one patients had stage I disease, 33 stage II, and 22 stage IIIA. Twenty-seven patients had poorly differentiated, 74 moderately differentiated, and 9 well-differentiated NSCLC; the remaining 16 patients were unspecified. Eighty patients were smokers, 39 former smokers, and the remaining seven never-smokers. One hundred and twenty-two patients underwent formal pulmonary lobectomy or more, with systematic ipsilateral mediastinal lymph node dissection; the four remaining patients underwent segmentectomy due to poor pulmonary reserve. Stages were determined after pathologic evaluation of resected specimens according to the International System for Staging Lung Cancer[30] (Table S1). None of the patients received adjuvant chemotherapy.
We validated the BRCA1 prognostic value in 58 stage IB-IIB NSCLC patients who had undergone surgical resection at the Azienda Ospedaliera Santa Maria (Terni, Italy) between February 1997 and December 2003, after obtaining approval from the institutional review board of Azienda Ospedaliera Santa Maria and patients' signed informed consent. Patient characteristics are shown in Table S1.
Gene expression analysis
Tumor samples from the 126 patients were obtained during surgery as blocks of 1cm3 and snap-frozen in liquid nitrogen. Tissues were stored in −80°C until total RNA was extracted with AllPrep kits (Qiagen, Valencia, CA). Only tumor samples containing more than 60% of tumor tissue on a microscopic section were eligible for further processing. The concentration of RNA was assessed in Nano-drop™ and the quality of obtained RNA was tested on agarose gel. First-strand cDNA was synthesized from 1 µg of total RNA using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). The nine genes examined are shown in Table 1. Quantitative RT-PCR reactions of each gene were done in an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems).
Table 1. Nine genes examined in this study.
| Official Symbol | Name | Accession number (transcript) (GEN ID) | Aliases |
| ERCC1 | excision repair cross-complementing rodent repair deficiency, complementation group 1 | NM_001983 | COFS4, UV20 |
| NM_202001 | |||
| (2067) | |||
| MZF1 | myeloid zinc finger 1 | NM_198055 | MZF-1, MZF1B, ZNF42, ZSCAN6, Zfp98 |
| NM_003422 | |||
| (7593) | |||
| Twist1 | twist homolog 1 (acrocephalosyndactyly 3; Saethre-Chotzen syndrome) (Drosophila) | NM_000474.3 | ACS3, BPES2, BPES3, SCS, TWIST |
| (7291) | |||
| RRM1 | ribonucleotide reductase M1 polypeptide | NM_001033 | R1, RIR1, RR1 |
| (6240) | |||
| TXN | thioredoxin | NM_003329 | DKFZp686B1993, MGC61975, TRX |
| (7295) | |||
| Tdp1 | tyrosyl-DNA phosphodiesterase | NM_018319 | FLJ11090, MGC104252 |
| NM_001008744 | |||
| (55775) | |||
| NFATC2 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | NM_173091 | RP5-1009H6.1, KIAA0611, NFAT1, NFATP |
| NM_012340 | |||
| (4773) | |||
| BRCA1 | breast cancer 1, early onset | NM_007294 | BRCAI, BRCC1, IRIS, PSCP, RNF53 |
| (672) | |||
| BUB1B | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | NM_001211 | BUB1beta, BUBR1, Bub1A, MAD3L, SSK1, hBUBR1 |
| (701) |
Relative gene expression values were calculated by the ΔΔCt method using the Sequence Detection System (SDS) 2.1 software (Applied Biosystems). The ΔΔCt method gives the amount of target gene normalized to an endogenous reference gene (ribosomal 18S RNA) and relative to a calibrator sample (reference for all samples; commercially available Normal Lung and Liver Human RNA (Stratagene, La Jolla, CA). Primers for the nine genes are listed in Table S2.
ERCC1, RRM1 and BRCA1 gene expression was assessed in formalin-fixed, paraffin-embedded surgical specimens from the 58 patients in the validation cohort. Using laser capture microdissection technique (Palm Microlaser, Oberlensheim, Germany) ensured a minimum of 80% of tumor tissue. After standard tissue sample deparaffinization using xylene and alcohols, samples were lysed in a tris-chloride, EDTA, sodium dodecyl sulphate (SDS) and proteinase K containing buffer. RNA was then extracted with phenol-chloroform-isoamyl alcohol followed by precipitation with isopropanol in the presence of glycogen and sodium acetate. RNA was resuspended in DEPC water (Ambion Inc, Austin TX, USA) and treated with DNAse I (Ambion Inc) to avoid DNA contamination. cDNA was synthesized using M-MLV retrotranscriptase enzyme. Template cDNA was added to Taqman Universal Master Mix (Applied Biosystems) in a 12.5-μl reaction with specific primers and probe for each gene. The primer and probe sets were identical to those used in the frozen specimens; the endogenous reference gene was β-actin. Quantification of gene expression was performed using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems).
Statistical analyses
Median values and ranges were derived for quantitative variables and mRNA gene expression. Qualitative variables were summarized by means of absolute frequencies and percentages. The Kruskal-Wallis test was used to check for normality. Differences in median mRNA expression levels between histological types were assessed by the U Mann-Whitney test. Spearman's rank-correlation coefficient (rho) was used to measure the correlations among gene expression levels. We made an a priori decision to classify mRNA gene-expression levels as high or low, using the minimum P value method modified by Lausen and Schumacher[31]. The Bonferroni method was used for the correction of the effect of multiple comparisons, and empirical P values for each gene were confirmed through 5000 permutation tests. When no optimal cut-off point was found, we used the sample median for the analysis of time to relapse and survival. Time to relapse and survival were calculated using Kaplan–Meier estimates and differences between curves were tested using the log-rank test. To choose an appropriate subset of genes for association to any clinical variable (histology, stage and grade), we performed a forward and backward Cox regression analysis. For all calculations, the tests performed were two-sided, significance was set at 5%, and the power was 80%. Analyses were performed using Statistical Package for the Social Sciences (SPSS) for Windows version 14 (SPSS Inc, Chicago, IL) and S-Plus 6.1 for Windows.
Results
The median values of each gene for the entire group of samples are shown in Table S3. Gene amplification was not successful in a minority of samples for every transcript analyzed. There were significant differences in expression according to histology for all genes except NFAT, with higher levels observed in squamous cell carcinomas than in adenocarcinomas (Table 2). There were no differences in gene expression according to stage (Table S4). A strong correlation was observed between expression levels of different genes, for example, between levels of TRX and RRM1 (rho = 0.52; P = 0.0003) and between ERCC1 and BRCA1 (rho = 0.62; P = 0.0001) (Table 3).
Table 2. Gene expression according to histology.
| Squamous cell carcinoma | Adenocarcinoma | P* | |
| Median (range) | Median (range) | ||
| ERCC1 | 1.41 (0.45–7.34) | 0.72 (0.23–2.45) | 0.0001 |
| MZF1 | 0.62 (0.06–6.72) | 0.25 (0.03–1.49) | 0.0001 |
| Twist | 10.37 (0.30–76.01) | 2.50 (0.14–19.16) | 0.0001 |
| RRM1 | 2 (0.6–6.9) | 1.2 (0.4–2.9) | 0.0001 |
| TRX | 2.13 (0.40–11.88) | 0.91 (0.31–7.94) | 0.0001 |
| Tdp1 | 1.7 (0.6–7.3) | 1.3 (0.1–2.6) | 0.02 |
| NFAT | 0.4 (0.1–2.3) | 0.5 (0.1–1.8) | 0.65 |
| BRCA1 | 4.26 (0.55–18.48) | 1.50 (0.09–8.08) | 0.0001 |
| BubR1 | 16.3 (1.4–90) | 7 (0.8–25) | 0.0001 |
Mann-Whitney U
Table 3. Correlation between expression levels of the nine genes examined.
| ERCC1 | MZF1 | Twist | RRM1 | TRX | Tdp1 | NFAT | BRCA1 | |
| MZF1 | 0.70 | |||||||
| P = 0.0001 | ||||||||
| Twist | 0.55 | 0.29 | ||||||
| P = 0.0001 | P = 0.001 | |||||||
| RRM1 | 0.33 | 0.27 | 0.24 | |||||
| P = 0.0001 | P = 0.004 | P = 0.01 | ||||||
| TRX | 0.39 | 0.20 | 0.25 | 0.52 | ||||
| P = 0.0001 | P = 0.03 | P = 0.005 | P = 0.0001 | |||||
| Tdp1 | 0.25 | 0.25 | 0.10 | 0.65 | 0.34 | |||
| P = 0.007 | P = 0.008 | P = 0.26 | P = 0.0001 | P = 0.0001 | ||||
| NFAT | 0.18 | 0.41 | −0.09 | −0.19 | −0.01 | −0.04 | ||
| P = 0.06 | P = 0.0001 | P = 0.36 | P = 0.04 | P = 0.88 | P = 0.66 | |||
| BRCA1 | 0.62 | 0.58 | 0.29 | 0.62 | 0.48 | 0.47 | 0.04 | |
| P = 0.0001 | P = 0.0001 | P = 0.001 | P = 0.0001 | P = 0.0001 | P = 0.0001 | P = 0.69 | ||
| BubR1 | 0.12 | 0.30 | 0.23 | 0.83 | 0.47 | 0.63 | −0.17 | 0.69 |
| P = 0.0001 | P = 0.001 | P = 0.01 | P = 0.0001 | P = 0.0001 | P = 0.0001 | P = 0.06 | P = 0.0001 |
With a median follow-up of 29.7 months (range, 1.7–65.9 months), overall event-free and median survival have not been reached. When event-free and median survival was analyzed according to expression levels of the nine genes, TRX and BRCA1 showed significant differences. Event-free survival for 21 patients with low TRX levels has not been reached, while it was 32 months (95%CI, ) for the remaining 93 patients with high levels (P = 0.02). For 77 patients with low levels of BRCA1, event-free survival has not been reached, while it was 22 months (95%CI, 14.9–29 months) for those with high levels (P = 0.04) (Table S5, Fig. 1). Event-free survival curves according to expression of the other seven genes are shown in Figure S1. Median survival for 24 patients with low TRX levels has not been reached, while it was 39 months for the remaining 101 patients with high levels (P = 0.03). For 83 patients with low levels of BRCA1, median survival has not been reached, while it was 29 months (95%CI, 22.2–35.7 months) for those with high levels (P = 0.04) (Table 4, Fig. 1). Median survival curves according to expression of the other seven genes are shown in Figure S2. However, when only stage I patients were examined, event-free survival was significantly different according to expression levels of MZF1 and BRCA1 (Table S6, Fig. S3), and median survival was significantly different according to expression levels of ERCC1, MZF1, Twist and BRCA1 (Table S7, Fig. S4).
Figure 1.
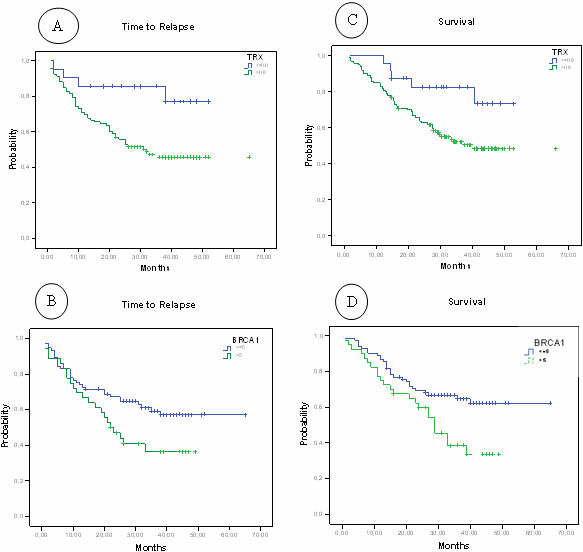
Event-free (A,B) and median (C,D) survival according to expression of TRX (A,C) and BRCA1 (B,D).
Table 4. Median survival according to gene expression.
| N* | Median Survival (months) | 95% CI | P | |
| ERCC1 | 0.89 | |||
| ≤1.24 | 61 | NR | - | |
| >1.24 | 62 | 39.5 | - | |
| MZF1 | 0.05 | |||
| ≤0.5 | 56 | NR | - | |
| >0.5 | 66 | 33 | 21.9–44.1 | |
| Twist | 0.39 | |||
| ≤7.75 | 61 | NR | - | |
| >7.75 | 61 | NR | - | |
| RRM1 | 0.11 | |||
| ≤1.65 | 61 | NR | - | |
| >1.65 | 60 | 33.9 | 24–43.9 | |
| TRX | 0.03 | |||
| ≤0.8 | 24 | NR | - | |
| >0.8 | 101 | 39 | - | |
| Tdp1 | 0.88 | |||
| ≤1.57 | 60 | NR | - | |
| >1.57 | 61 | 41.3 | - | |
| NFAT | 0.51 | |||
| ≤0.46 | 61 | NR | - | |
| >0.46 | 61 | 33.9 | - | |
| BRCA1 | 0.01 | |||
| ≤5 | 83 | NR | - | |
| >5 | 40 | 29 | 22.2–35.7 | |
| BubR1 | 0.41 | |||
| ≤12.28 | 61 | NR | - | |
| >12.28 | 61 | 36.7 | - |
Survival data is not available for some patients. Gene amplification was not successfully performed in all samples for all genes.
NR = not reached
The Cox proportional hazards model selected pathological stage IIIA and BRCA1 expression as independent prognostic factors for survival. The hazard ratio (HR) was 7.91 (95%CI, 2.27-27.54; P = 0.001) for stage IIIA and 1.98 (95%CI, 1.11-6); P = 0.02) for BRCA1 expression (Table S8).
Validation of BRCA1
The median follow-up of the 58 patients in the validation cohort was 40 months. According to the Cox proportional hazards model, the HR for patients with high levels of BRCA1 was 2.4 (95%CI, 1.01-5.92; P = 0.04). There were no stage IIIA patients in this cohort.
Discussion
Gene expression signatures have been shown to predict outcome in resected stage I NSCLC[5], [6], However, the use of microarrays is limited due to the need for fresh-frozen tissue. RT-QPCR involving a small number of genes offers a practical alternative, allowing for accurate and reproducible quantification of results for RNA obtained from small amounts of paraffin-embedded specimens. The results of RT-QPCR performed on five[7] and eight[32] genes correlated with outcomes of NSCLC[7] and lung adenocarcinoma[32] patients. We have examined the expression of ERCC1, BRCA1, BubR1, MZF1, RRM1, TRX1 and Tdp1, involved in DNA repair pathways, and of Twist and NFAT , related to metastasis formation. In the multivariate model, only BRCA1 and stage IIIA were identified as independent prognostic variables. In an independent validation cohort of 58 stage IB-IIB NSCLC patients, BRCA1 was confirmed as the only independent prognostic marker.
Patients whose tumors had high BRCA1 expression had significantly worse survival and should be candidates for adjuvant chemotherapy. In vitro studies have shown that BRCA1 can regulate differential sensitivity to different classes of chemotherapy agents[33]. The absence of BRCA1 results in high sensitivity to cisplatin, whereas its presence increases sensitivity to antimicrotubule agents[33]. Therefore, we believe that patients with the highest expression levels should receive antimicrotubule, non-platinum-based chemotherapy. We have carried out a pilot study of customized adjuvant chemotherapy based on BRCA1 mRNA levels in 88 completely resected stage II-IIIA NSCLC patients, where those with the highest expression levels received adjuvant docetaxel and those with lower levels received cisplatin-based chemotherapy. The interim analysis shows that event-free survival is similar in both groups. These findings support our previous findings in stage II-IIIA patients who received neoadjuvant gemcitabine/cisplatin, where those with the highest BRCA1 levels had a dismal survival of 12 months[11].
No differences in expression levels of any of the nine genes were observed according to stage or tumor size (<4 vs >4 cms). However, of all the nine genes examined, only BRCA1 showed a trend towards influencing survival according to tumor size. In stage I NSCLC patients, survival has been inversely correlated with tumor size[34]. In the present study, the univariate survival analysis showed that in addition to BRCA1, ERCC1 and MZF1 significantly influenced survival in stage I (Table S7, Fig. S4). These findings highlight the potential role of ERCC1 and MZF1, which are highly correlated with BRCA1, as strong prognostic markers in stage I NSCLC. Not unexpectedly, however, considering the high correlation between the expression levels of these three genes (Table 3), when all three genes were combined, no further improvement over the prognostic value of BRCA1 alone was observed.
Although the mechanisms by which some of the nine genes examined affect patient prognosis is not very clear, overexpression of ERCC1 and RRM1 seems to be oncogene-driven[35], [36], [37], [38], [39]. BRCA1 methylation and abrogation of BRCA1 mRNA has been found in sporadic breast cancers[40] but very rarely in NSCLC[41]. In some sporadic breast cancers, the poor outcome associated with BRCA1 methylation and low levels of expression could be explained by MYC amplification[42].
Other studies, using the monoclonal antibody 8F1[43] have reported that the presence of ERCC1 protein is a prognostic marker of survival in early NSCLC and a predictor of outcome to adjuvant cisplatin-based chemotherapy; however, in a prior study in gastric cancer[44], it was unclear whether the poor clinical response of patients whose tumors had high pretreatment mRNA levels of ERCC1 resulted from tumor cell resistance to cisplatin-based chemotherapy or from a more aggressive tumor biology. Moreover, in ERCC1-positive normal human fibroblasts and cells from patients with inherited mutations in ERCC1, ERCC1 is not the principal antigen recognized by the 8F1 antibody on immunostaining[45]. Furthermore, in another study, ERCC1 protein status did not correlate with survival in stage IV NSCLC[46], while in a trial of customized cisplatin based on ERCC1 mRNA expression, response rate was 39% in the control arm and 50% in the customized arm (P = 0.02)[47].
In summary, our study indicates that firstly, BRCA1 is closely related to ERCC1, RRM1 and other genes like MZF1, but stands out as the most significant prognostic marker of relapse. We hypothesize that patients with high BRCA1 levels will benefit from antimicrotubule-based–but not cisplatin-based–chemotherapy. Secondly, high levels of these transcripts confer a higher risk of relapse, in contrast to what has been reported by other investigators, which highlights the need for further research in this area to elucidate the predictive role of these NER-related genes and to correctly customize treatment (Fig. S5). Although the population in our study was skewed to male smokers with squamous cell carcinoma, our results warrant further investigation to confirm their applicability to other histological subsets of NSCLC, In order to shed further light on these issues, we are planning to examine BRCA1, ERCC1, MZF1 and RRM1 expression in 200 tumor specimens from the ANITA study[3] and in 620 patients included in the Spanish Lung Cancer Group NATCH trial of neoadjuvant vs adjuvant chemotherapy vs surgery alone.
Supporting Information
Event-free survival according to the expression of ERCC1 (A), MZF1 (B), Twist (C), RRM1 (D), Tdp1 (E), NFAT (F), and BubR1 (G)
(0.09 MB TIF)
Median survival according to the expression of ERCC1 (A), MZF1 (B), Twist (C), RRM1 (D), Tdp1 (E), NFAT (F), and BubR1 (G)
(0.09 MB TIF)
Event-free survival curves for stage I patients according to gene expression levels of the nine genes examined
(0.10 MB TIF)
Median survival curves for stage I patients according to gene expression levels of the nine genes examined
(0.10 MB TIF)
Contradictory findings leading to opposed strategies of customizing adjuvant chemotherapy. Olaussen et al (NEJM 2006;355:983-991) report that the lack of ERCC1protein implies a higher risk of relapse and a greater sensitivity to cisplatin-based chemotherapy. (Cisplatin sensitivity based on lack of ERCC1 expression has been demonstrated in preclinical and clinical studies.) Our findings indicate that a higher risk of relapse is related to high levels of several transcripts, including ERCC1. These patients could be resistant to cisplatin and sensitive to taxanes or other antimicrotubule drugs
(0.13 MB TIF)
Patient characteristics for principal cohort (N = 126) and for validation cohort (N = 58)
(0.04 MB DOC)
Primers and probes for the nine genes examined
(0.03 MB DOC)
Relative gene expression values
(0.03 MB DOC)
Gene expression according to disease stage
(0.04 MB DOC)
Event-free survival according to gene expression levels
(0.06 MB DOC)
Event-free survival in stage I patients according to gene expression levels
(0.06 MB DOC)
Median survival for stage I patients according to gene expression levels
(0.06 MB DOC)
Multivariate Cox model for survival, showing a greater risk of death for patients with high levels of BRCA1 and for those with stage IIIA disease
(0.03 MB DOC)
Further details on the nine genes examined
(0.09 MB DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The research reported here was funded by the Spanish Lung Cancer Group and the Medical University of Gdansk. It was partially funded by the Spanish Ministry of Health grant FIS 05/1621. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, et al. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol. 2007;18:581–592. doi: 10.1093/annonc/mdl498. [DOI] [PubMed] [Google Scholar]
- 2.Strauss GM. Adjuvant chemotherapy of lung cancer: methodologic issues and therapeutic advances. Hematol Oncol Clin North Am. 2005;19:263–281, vi. doi: 10.1016/j.hoc.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Douillard JY, Rosell R, De Lena M, Carpagnano F, Ramlau R, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol. 2006;7:719–727. doi: 10.1016/S1470-2045(06)70804-X. [DOI] [PubMed] [Google Scholar]
- 4.Parmigiani G, Garrett-Mayer ES, Anbazhagan R, Gabrielson E. A cross-study comparison of gene expression studies for the molecular classification of lung cancer. Clin Cancer Res. 2004;10:2922–2927. doi: 10.1158/1078-0432.ccr-03-0490. [DOI] [PubMed] [Google Scholar]
- 5.Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y, Lemon W, Liu PY, Yi Y, Morrison C, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 8.Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet. 2007 doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- 9.Furuta T, Ueda T, Aune G, Sarasin A, Kraemer KH, et al. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res. 2002;62:4899–4902. [PubMed] [Google Scholar]
- 10.Lord RV, Brabender J, Gandara D, Alberola V, Camps C, et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res. 2002;8:2286–2291. [PubMed] [Google Scholar]
- 11.Taron M, Rosell R, Felip E, Mendez P, Souglakos J, et al. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet. 2004;13:2443–2449. doi: 10.1093/hmg/ddh260. [DOI] [PubMed] [Google Scholar]
- 12.Bae I, Rih JK, Kim HJ, Kang HJ, Haddad B, et al. BRCA1 regulates gene expression for orderly mitotic progression. Cell Cycle. 2005;4:1641–1666. doi: 10.4161/cc.4.11.2152. [DOI] [PubMed] [Google Scholar]
- 13.Chabalier C, Lamare C, Racca C, Privat M, Valette A, et al. BRCA1 downregulation leads to premature inactivation of spindle checkpoint and confers paclitaxel resistance. Cell Cycle. 2006;5:1001–1007. doi: 10.4161/cc.5.9.2726. [DOI] [PubMed] [Google Scholar]
- 14.Shichiri M, Yoshinaga K, Hisatomi H, Sugihara K, Hirata Y. Genetic and epigenetic inactivation of mitotic checkpoint genes hBUB1 and hBUBR1 and their relationship to survival. Cancer Res. 2002;62:13–17. [PubMed] [Google Scholar]
- 15.Hromas R, Collins SJ, Hickstein D, Raskind W, Deaven LL, et al. A retinoic acid-responsive human zinc finger gene, MZF-1, preferentially expressed in myeloid cells. J Biol Chem. 1991;266:14183–14187. [PubMed] [Google Scholar]
- 16.Yan QW, Reed E, Zhong XS, Thornton K, Guo Y, et al. MZF1 possesses a repressively regulatory function in ERCC1 expression. Biochem Pharmacol. 2006;71:761–771. doi: 10.1016/j.bcp.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 17.Le Mee S, Fromigue O, Marie PJ. Sp1/Sp3 and the myeloid zinc finger gene MZF1 regulate the human N-cadherin promoter in osteoblasts. Exp Cell Res. 2005;302:129–142. doi: 10.1016/j.yexcr.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Rosell R, Scagliotti G, Danenberg KD, Lord RV, Bepler G, et al. Transcripts in pretreatment biopsies from a three-arm randomized trial in metastatic non-small-cell lung cancer. Oncogene. 2003;22:3548–3553. doi: 10.1038/sj.onc.1206419. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Z, Chen T, Li X, Haura E, Sharma A, et al. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med. 2007;356:800–808. doi: 10.1056/NEJMoa065411. [DOI] [PubMed] [Google Scholar]
- 20.Rosell R, Felip E, Taron M, Majo J, Mendez P, et al. Gene expression as a predictive marker of outcome in stage IIB-IIIA-IIIB non-small cell lung cancer after induction gemcitabine-based chemotherapy followed by resectional surgery. Clin Cancer Res. 2004;10:4215s–4219s. doi: 10.1158/1078-0432.CCR-040006. [DOI] [PubMed] [Google Scholar]
- 21.Kakolyris S, Giatromanolaki A, Koukourakis M, Powis G, Souglakos J, et al. Thioredoxin expression is associated with lymph node status and prognosis in early operable non-small cell lung cancer. Clin Cancer Res. 2001;7:3087–3091. [PubMed] [Google Scholar]
- 22.Yoshida T, Nakamura H, Masutani H, Yodoi J. The involvement of thioredoxin and thioredoxin binding protein-2 on cellular proliferation and aging process. Ann N Y Acad Sci. 2005;1055:1–12. doi: 10.1196/annals.1323.002. [DOI] [PubMed] [Google Scholar]
- 23.Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, et al. SCAN1 mutant Tdp1 accumulates the enzyme–DNA intermediate and causes camptothecin hypersensitivity. Embo J. 2005;24:2224–2233. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barthelmes HU, Habermeyer M, Christensen MO, Mielke C, Interthal H, et al. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J Biol Chem. 2004;279:55618–55625. doi: 10.1074/jbc.M405042200. [DOI] [PubMed] [Google Scholar]
- 25.Liu C, Zhou S, Begum S, Sidransky D, Westra WH, et al. Increased expression and activity of repair genes TDP1 and XPF in non-small cell lung cancer. Lung Cancer. 2007;55:303–311. doi: 10.1016/j.lungcan.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Alexander NR, Tran NL, Rekapally H, Summers CE, Glackin C, et al. N-cadherin gene expression in prostate carcinoma is modulated by integrin-dependent nuclear translocation of Twist1. Cancer Res. 2006;66:3365–3369. doi: 10.1158/0008-5472.CAN-05-3401. [DOI] [PubMed] [Google Scholar]
- 28.Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, et al. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell. 2005;20:539–550. doi: 10.1016/j.molcel.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 29.Jauliac S, Lopez-Rodriguez C, Shaw LM, Brown LF, Rao A, et al. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002;4:540–544. doi: 10.1038/ncb816. [DOI] [PubMed] [Google Scholar]
- 30.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 31.Lausen B, Schumacher M. Maximally selected rank statistics. Biometrics. 1992;48:73–85. [Google Scholar]
- 32.Endoh H, Tomida S, Yatabe Y, Konishi H, Osada H, et al. Prognostic model of pulmonary adenocarcinoma by expression profiling of eight genes as determined by quantitative real-time reverse transcriptase polymerase chain reaction. J Clin Oncol. 2004;22:811–819. doi: 10.1200/JCO.2004.04.109. [DOI] [PubMed] [Google Scholar]
- 33.Quinn JE, Kennedy RD, Mullan PB, Gilmore PM, Carty M, et al. BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res. 2003;63:6221–6228. [PubMed] [Google Scholar]
- 34.Wisnivesky JP, Yankelevitz D, Henschke CI. The effect of tumor size on curability of stage I non-small cell lung cancers. Chest. 2004;126:761–765. doi: 10.1378/chest.126.3.761. [DOI] [PubMed] [Google Scholar]
- 35.Potapova O, Haghighi A, Bost F, Liu C, Birrer MJ, et al. The Jun kinase/stress-activated protein kinase pathway functions to regulate DNA repair and inhibition of the pathway sensitizes tumor cells to cisplatin. J Biol Chem. 1997;272:14041–14044. doi: 10.1074/jbc.272.22.14041. [DOI] [PubMed] [Google Scholar]
- 36.Zhao R, Rabo YB, Egyhazi S, Andersson A, Edgren MR, et al. Apoptosis and c-jun induction by cisplatin in a human melanoma cell line and a drug-resistant daughter cell line. Anticancer Drugs. 1995;6:657–668. doi: 10.1097/00001813-199510000-00005. [DOI] [PubMed] [Google Scholar]
- 37.Moorehead RA, Singh G. Influence of the proto-oncogene c-fos on cisplatin sensitivity. Biochem Pharmacol. 2000;59:337–345. doi: 10.1016/s0006-2952(99)00333-0. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura H. Thioredoxin as a key molecule in redox signaling. Antioxid Redox Signal. 2004;6:15–17. doi: 10.1089/152308604771978309. [DOI] [PubMed] [Google Scholar]
- 39.Welsh SJ, Bellamy WT, Briehl MM, Powis G. The redox protein thioredoxin-1 (Trx-1) increases hypoxia-inducible factor 1alpha protein expression: Trx-1 overexpression results in increased vascular endothelial growth factor production and enhanced tumor angiogenesis. Cancer Res. 2002;62:5089–5095. [PubMed] [Google Scholar]
- 40.Rice JC, Massey-Brown KS, Futscher BW. Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene. 1998;17:1807–1812. doi: 10.1038/sj.onc.1202086. [DOI] [PubMed] [Google Scholar]
- 41.Marsit CJ, Liu M, Nelson HH, Posner M, Suzuki M, et al. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene. 2004;23:1000–1004. doi: 10.1038/sj.onc.1207256. [DOI] [PubMed] [Google Scholar]
- 42.Grushko TA, Dignam JJ, Das S, Blackwood AM, Perou CM, et al. MYC is amplified in BRCA1-associated breast cancers. Clin Cancer Res. 2004;10:499–507. doi: 10.1158/1078-0432.ccr-0976-03. [DOI] [PubMed] [Google Scholar]
- 43.Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 44.Metzger R, Leichman CG, Danenberg KD, Danenberg PV, Lenz HJ, et al. ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. J Clin Oncol. 1998;16:309–316. doi: 10.1200/JCO.1998.16.1.309. [DOI] [PubMed] [Google Scholar]
- 45.Niedernhofer LJ, Bhagwat N, Wood RD. ERCC1 and non-small-cell lung cancer. N Engl J Med. 2007;356:2538–2540; author reply 2540–2531. doi: 10.1056/NEJMc070742. [DOI] [PubMed] [Google Scholar]
- 46.Wachters FM, Wong LS, Timens W, Kampinga HH, Groen HJ. ERCC1, hRad51, and BRCA1 protein expression in relation to tumour response and survival of stage III/IV NSCLC patients treated with chemotherapy. Lung Cancer. 2005;50:211–219. doi: 10.1016/j.lungcan.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 47.Cobo M, Isla D, Massuti B, Montes A, Sanchez JM, et al. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol. 2007;25:2747–2754. doi: 10.1200/JCO.2006.09.7915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Event-free survival according to the expression of ERCC1 (A), MZF1 (B), Twist (C), RRM1 (D), Tdp1 (E), NFAT (F), and BubR1 (G)
(0.09 MB TIF)
Median survival according to the expression of ERCC1 (A), MZF1 (B), Twist (C), RRM1 (D), Tdp1 (E), NFAT (F), and BubR1 (G)
(0.09 MB TIF)
Event-free survival curves for stage I patients according to gene expression levels of the nine genes examined
(0.10 MB TIF)
Median survival curves for stage I patients according to gene expression levels of the nine genes examined
(0.10 MB TIF)
Contradictory findings leading to opposed strategies of customizing adjuvant chemotherapy. Olaussen et al (NEJM 2006;355:983-991) report that the lack of ERCC1protein implies a higher risk of relapse and a greater sensitivity to cisplatin-based chemotherapy. (Cisplatin sensitivity based on lack of ERCC1 expression has been demonstrated in preclinical and clinical studies.) Our findings indicate that a higher risk of relapse is related to high levels of several transcripts, including ERCC1. These patients could be resistant to cisplatin and sensitive to taxanes or other antimicrotubule drugs
(0.13 MB TIF)
Patient characteristics for principal cohort (N = 126) and for validation cohort (N = 58)
(0.04 MB DOC)
Primers and probes for the nine genes examined
(0.03 MB DOC)
Relative gene expression values
(0.03 MB DOC)
Gene expression according to disease stage
(0.04 MB DOC)
Event-free survival according to gene expression levels
(0.06 MB DOC)
Event-free survival in stage I patients according to gene expression levels
(0.06 MB DOC)
Median survival for stage I patients according to gene expression levels
(0.06 MB DOC)
Multivariate Cox model for survival, showing a greater risk of death for patients with high levels of BRCA1 and for those with stage IIIA disease
(0.03 MB DOC)
Further details on the nine genes examined
(0.09 MB DOC)


