Abstract
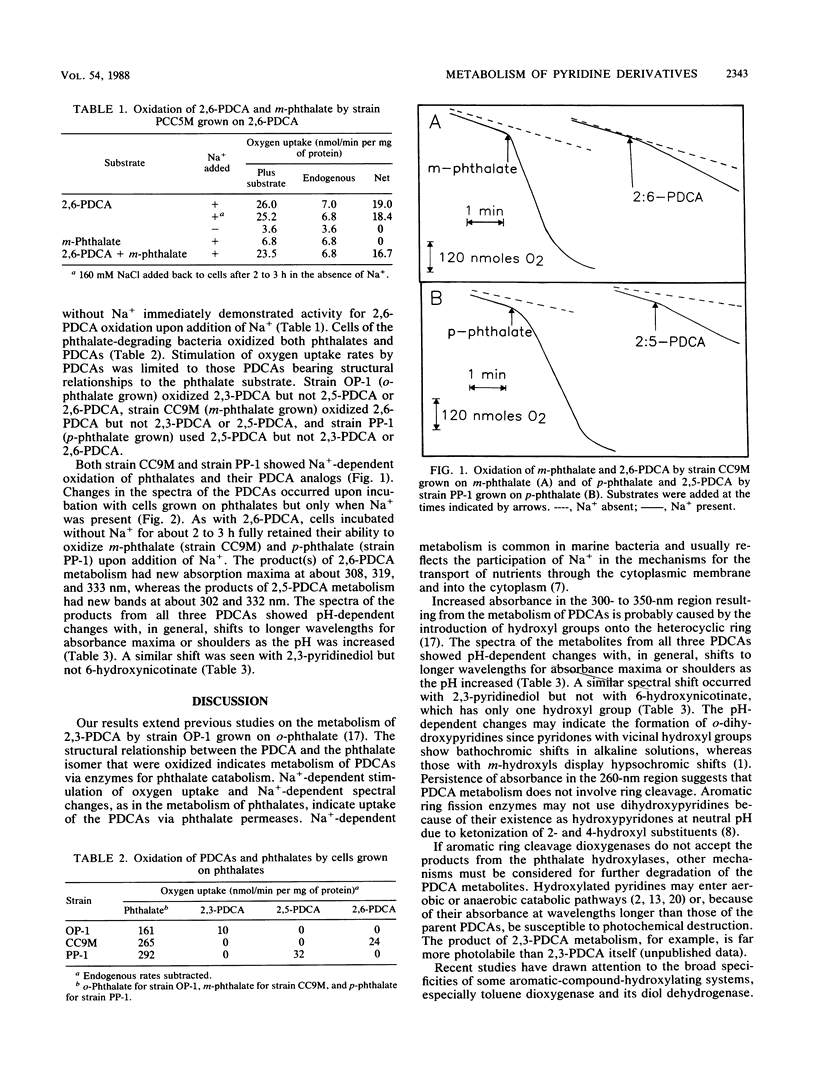
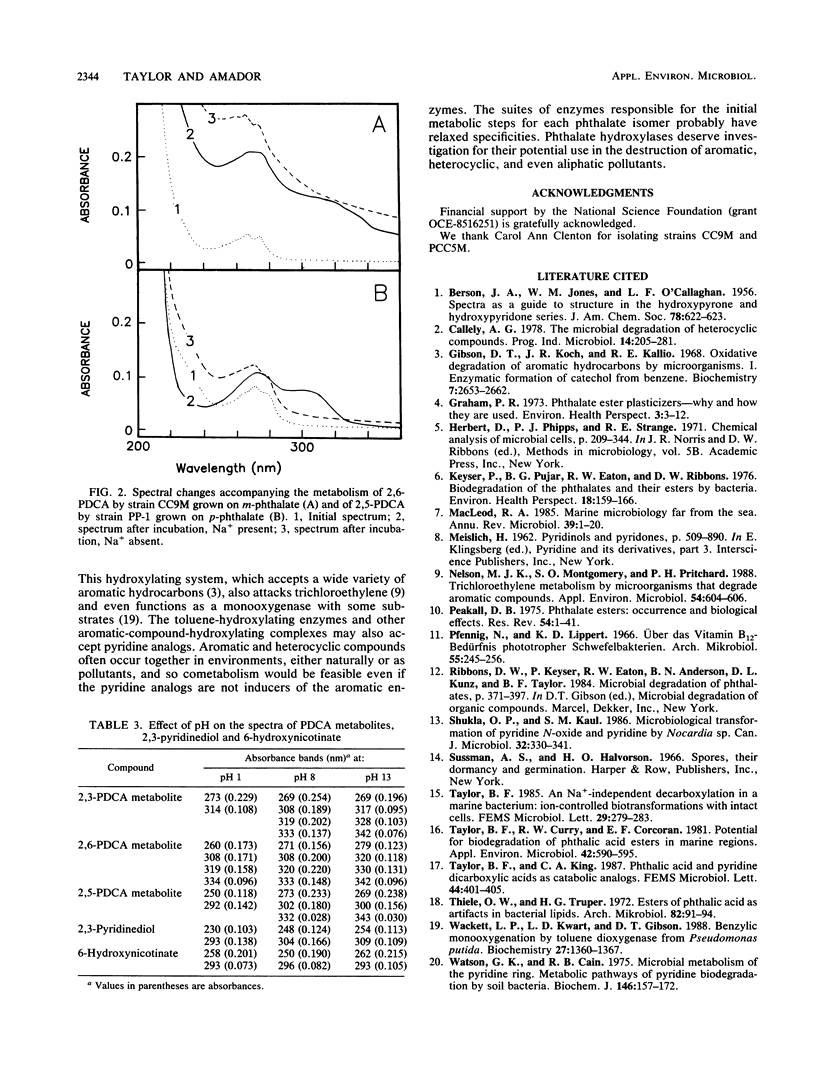
Bacteria were isolated from marine sediments that grew aerobically on m-phthalate, p-phthalate, or dipicolinate (2,6-pyridine dicarboxylate [2,6-PDCA]). Strain OP-1, which grew on o-phthalate and was previously obtained from a marine source, was also studied. Intact cells of each organism demonstrated Na+-dependent oxidation of their growth substrates. Strain PCC5M grew on dipicolinate but did not metabolize m-phthalate. The phthalate degraders, however, demonstrated Na+-dependent metabolism of the appropriate PDCA analogs. 2,6-PDCA was transformed by strain CC9M when this strain was grown on m-phthalate, 2,5-PDCA was metabolized by strain PP-1 grown on p-phthalate, and 2,3-PDCA (quinolinate) was oxidized by strain OP-1 grown on o-phthalate. Spectral changes accompanying the Na+-dependent transformations of the PDCA analogs suggest the formation of hydroxylated compounds. Metabolism probably occurred via phthalate hydroxylases; this is a previously unrecognized route for the environmental transformation of pyridine compounds. Hydroxylated products may feed into known pathways for the catabolism of pyridines or be photochemically degraded because of their absorbance in the solar actinic range (wavelengths > 300 nm). The results reinforce recent evidence for the broad potential of aromatic hydroxylase systems for the destruction of pollutants.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gibson D. T., Koch J. R., Kallio R. E. Oxidative degradation of aromatic hydrocarbons by microorganisms. I. Enzymatic formation of catechol from benzene. Biochemistry. 1968 Jul;7(7):2653–2662. doi: 10.1021/bi00847a031. [DOI] [PubMed] [Google Scholar]
- Graham P. R. Phthalate ester plasticizers--why and how they are used. Environ Health Perspect. 1973 Jan;3:3–12. doi: 10.1289/ehp.73033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser P., Pujar B. G., Eaton R. W., Ribbons D. W. Biodegradation of the phthalates and their esters by bacteria. Environ Health Perspect. 1976 Dec;18:159–166. doi: 10.1289/ehp.7618159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod R. A. Marine microbiology far from the sea. Annu Rev Microbiol. 1985;39:1–20. doi: 10.1146/annurev.mi.39.100185.000245. [DOI] [PubMed] [Google Scholar]
- Nelson M. J., Montgomery S. O., Pritchard P. H. Trichloroethylene metabolism by microorganisms that degrade aromatic compounds. Appl Environ Microbiol. 1988 Feb;54(2):604–606. doi: 10.1128/aem.54.2.604-606.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall D. B. Phthalate esters: Occurrence and biological effects. Residue Rev. 1975;54:1–41. doi: 10.1007/978-1-4612-9857-1_1. [DOI] [PubMed] [Google Scholar]
- Taylor B. F., Curry R. W., Corcoran E. F. Potential for biodegradation of phthalic Acid esters in marine regions. Appl Environ Microbiol. 1981 Oct;42(4):590–595. doi: 10.1128/aem.42.4.590-595.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele O. W., Trüper H. G. Esters of phthalic acid as artifacts in bacterial lipids. Arch Mikrobiol. 1972;82(1):91–94. doi: 10.1007/BF00424932. [DOI] [PubMed] [Google Scholar]
- Wackett L. P., Kwart L. D., Gibson D. T. Benzylic monooxygenation catalyzed by toluene dioxygenase from Pseudomonas putida. Biochemistry. 1988 Feb 23;27(4):1360–1367. doi: 10.1021/bi00404a041. [DOI] [PubMed] [Google Scholar]
- Watson G. K., Cain R. B. Microbial metabolism of the pyridine ring. Metabolic pathways of pyridine biodegradation by soil bacteria. Biochem J. 1975 Jan;146(1):157–172. doi: 10.1042/bj1460157. [DOI] [PMC free article] [PubMed] [Google Scholar]


