Abstract
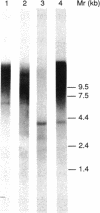
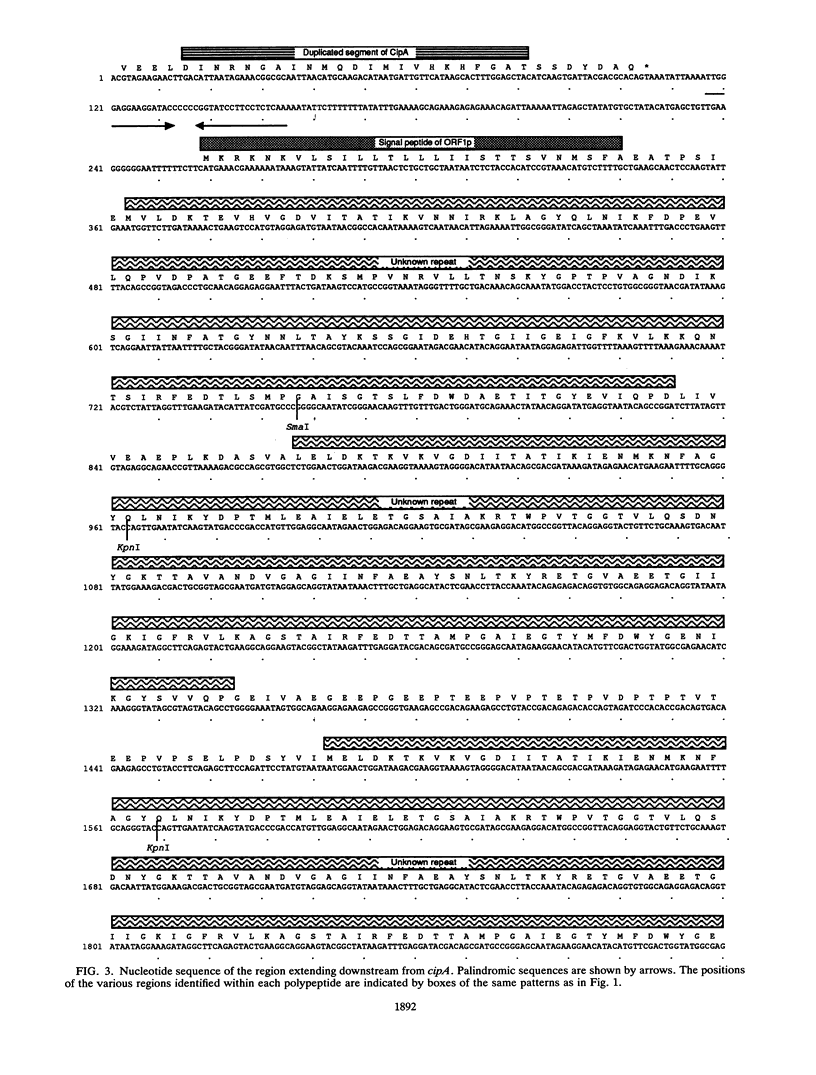
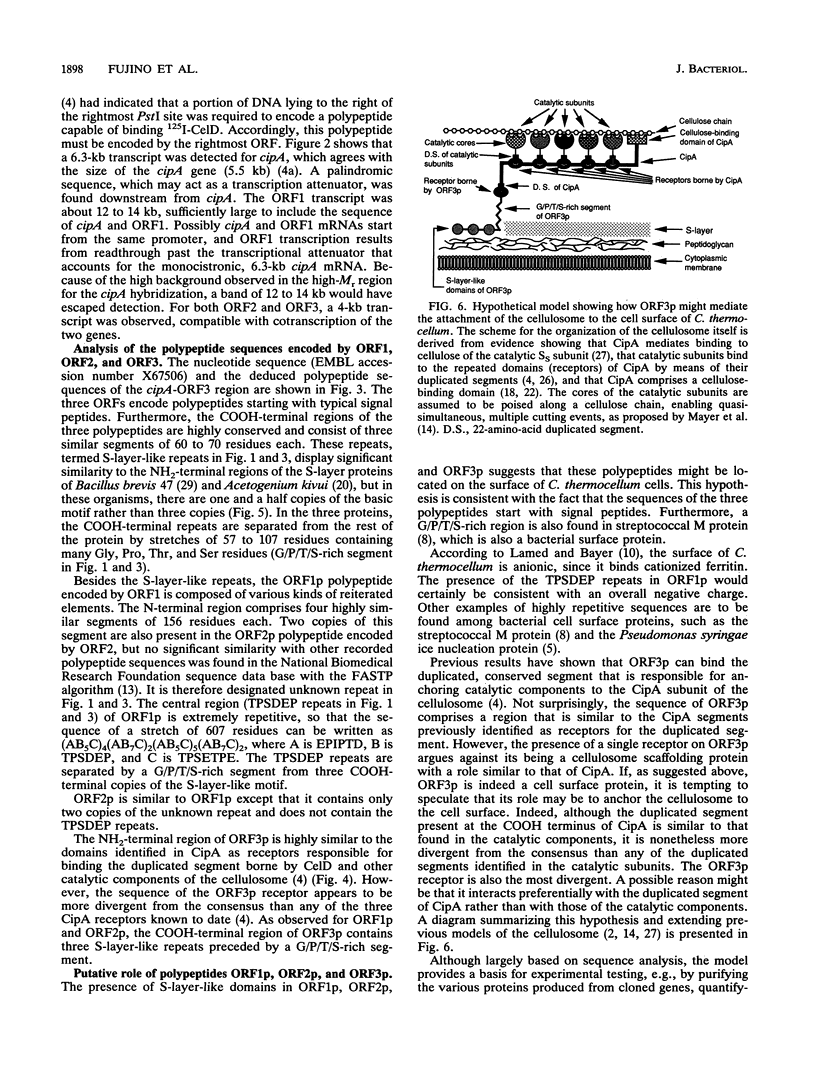
The nucleotide sequence was determined for a 9.4-kb region of Clostridium thermocellum DNA extending from the 3' end of the gene (now termed cipA), encoding the S1/SL component of the cellulosome. Three open reading frames (ORFs) belonging to two operons were detected. They encoded polypeptides of 1,664, 688, and 447 residues, termed ORF1p, ORF2p, and ORF3p, respectively. The COOH-terminal regions of the three polypeptides were highly similar and contained three reiterated segments of 60 to 70 residues each. Similar segments have been found at the NH2 terminus of the S-layer proteins of Bacillus brevis and Acetogenium kivui, suggesting that ORF1p, ORF2p, and ORF3p might also be located on the cell surface. Otherwise, the sequence of ORF1p and ORF2p gave little clue concerning their potential function. However, the NH2-terminal region of ORF3p was similar to the reiterated domains previously identified in CipA as receptors involved in binding the duplicated segment of 22 amino acids present in catalytic subunits of the cellulosome. Indeed, it was found previously that ORF3p binds 125I-labeled endoglucanase CelD containing the duplicated segment (T. Fujino, P. Béguin, and J.-P. Aubert, FEMS Microbiol. Lett. 94:165-170, 1992). These findings suggest that ORF3p might serve as an anchoring factor for the cellulosome on the cell surface by binding the duplicated segment that is present at the COOH end of CipA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Béguin P. Detection of cellulase activity in polyacrylamide gels using Congo red-stained agar replicas. Anal Biochem. 1983 Jun;131(2):333–336. doi: 10.1016/0003-2697(83)90178-1. [DOI] [PubMed] [Google Scholar]
- Béguin P., Millet J., Aubert J. P. Cellulose degradation by Clostridium thermocellum: from manure to molecular biology. FEMS Microbiol Lett. 1992 Dec 15;100(1-3):523–528. doi: 10.1111/j.1574-6968.1992.tb14087.x. [DOI] [PubMed] [Google Scholar]
- Coughlan M. P., Hon-Nami K., Hon-Nami H., Ljungdahl L. G., Paulin J. J., Rigsby W. E. The cellulolytic enzyme complex of Clostridium thermocellum is very large. Biochem Biophys Res Commun. 1985 Jul 31;130(2):904–909. doi: 10.1016/0006-291x(85)90502-9. [DOI] [PubMed] [Google Scholar]
- Fujino T., Béguin P., Aubert J. P. Cloning of a Clostridium thermocellum DNA fragment encoding polypeptides that bind the catalytic components of the cellulosome. FEMS Microbiol Lett. 1992 Jul 1;73(1-2):165–170. doi: 10.1016/0378-1097(92)90602-k. [DOI] [PubMed] [Google Scholar]
- Grépinet O., Chebrou M. C., Béguin P. Purification of Clostridium thermocellum xylanase Z expressed in Escherichia coli and identification of the corresponding product in the culture medium of C. thermocellum. J Bacteriol. 1988 Oct;170(10):4576–4581. doi: 10.1128/jb.170.10.4576-4581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hollingshead S. K., Fischetti V. A., Scott J. R. Complete nucleotide sequence of type 6 M protein of the group A Streptococcus. Repetitive structure and membrane anchor. J Biol Chem. 1986 Feb 5;261(4):1677–1686. [PubMed] [Google Scholar]
- Kohring S., Wiegel J., Mayer F. Subunit Composition and Glycosidic Activities of the Cellulase Complex from Clostridium thermocellum JW20. Appl Environ Microbiol. 1990 Dec;56(12):3798–3804. doi: 10.1128/aem.56.12.3798-3804.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamed R., Setter E., Bayer E. A. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J Bacteriol. 1983 Nov;156(2):828–836. doi: 10.1128/jb.156.2.828-836.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mayer F., Coughlan M. P., Mori Y., Ljungdahl L. G. Macromolecular Organization of the Cellulolytic Enzyme Complex of Clostridium thermocellum as Revealed by Electron Microscopy. Appl Environ Microbiol. 1987 Dec;53(12):2785–2792. doi: 10.1128/aem.53.12.2785-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Mishra S., Béguin P., Aubert J. P. Transcription of Clostridium thermocellum endoglucanase genes celF and celD. J Bacteriol. 1991 Jan;173(1):80–85. doi: 10.1128/jb.173.1.80-85.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morag E., Bayer E. A., Lamed R. Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocellum to cellulose-degrading enzymes. J Bacteriol. 1990 Oct;172(10):6098–6105. doi: 10.1128/jb.172.10.6098-6105.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morag E., Halevy I., Bayer E. A., Lamed R. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J Bacteriol. 1991 Jul;173(13):4155–4162. doi: 10.1128/jb.173.13.4155-4162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J., Peters M., Lottspeich F., Baumeister W. S-layer protein gene of Acetogenium kivui: cloning and expression in Escherichia coli and determination of the nucleotide sequence. J Bacteriol. 1989 Nov;171(11):6307–6315. doi: 10.1128/jb.171.11.6307-6315.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniec M. P., Kobayashi T., Fauth U., Gerngross U. T., Demain A. L. Cloning and expression of a Clostridium thermocellum DNA fragment that encodes a protein related to cellulosome component SL. Appl Biochem Biotechnol. 1991 Nov;31(2):119–134. doi: 10.1007/BF02921783. [DOI] [PubMed] [Google Scholar]
- Salamitou S., Tokatlidis K., Béguin P., Aubert J. P. Involvement of separate domains of the cellulosomal protein S1 of Clostridium thermocellum in binding to cellulose and in anchoring of catalytic subunits to the cellulosome. FEBS Lett. 1992 Jun 8;304(1):89–92. doi: 10.1016/0014-5793(92)80595-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tailliez P., Girard H., Millet J., Beguin P. Enhanced Cellulose Fermentation by an Asporogenous and Ethanol-Tolerant Mutant of Clostridium thermocellum. Appl Environ Microbiol. 1989 Jan;55(1):207–211. doi: 10.1128/aem.55.1.207-211.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA transferred or dotted nitrocellulose paper. Methods Enzymol. 1983;100:255–266. doi: 10.1016/0076-6879(83)00060-9. [DOI] [PubMed] [Google Scholar]
- Tokatlidis K., Salamitou S., Béguin P., Dhurjati P., Aubert J. P. Interaction of the duplicated segment carried by Clostridium thermocellum cellulases with cellulosome components. FEBS Lett. 1991 Oct 21;291(2):185–188. doi: 10.1016/0014-5793(91)81279-h. [DOI] [PubMed] [Google Scholar]
- Yamagata H., Adachi T., Tsuboi A., Takao M., Sasaki T., Tsukagoshi N., Udaka S. Cloning and characterization of the 5' region of the cell wall protein gene operon in Bacillus brevis 47. J Bacteriol. 1987 Mar;169(3):1239–1245. doi: 10.1128/jb.169.3.1239-1245.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]