Abstract
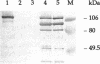
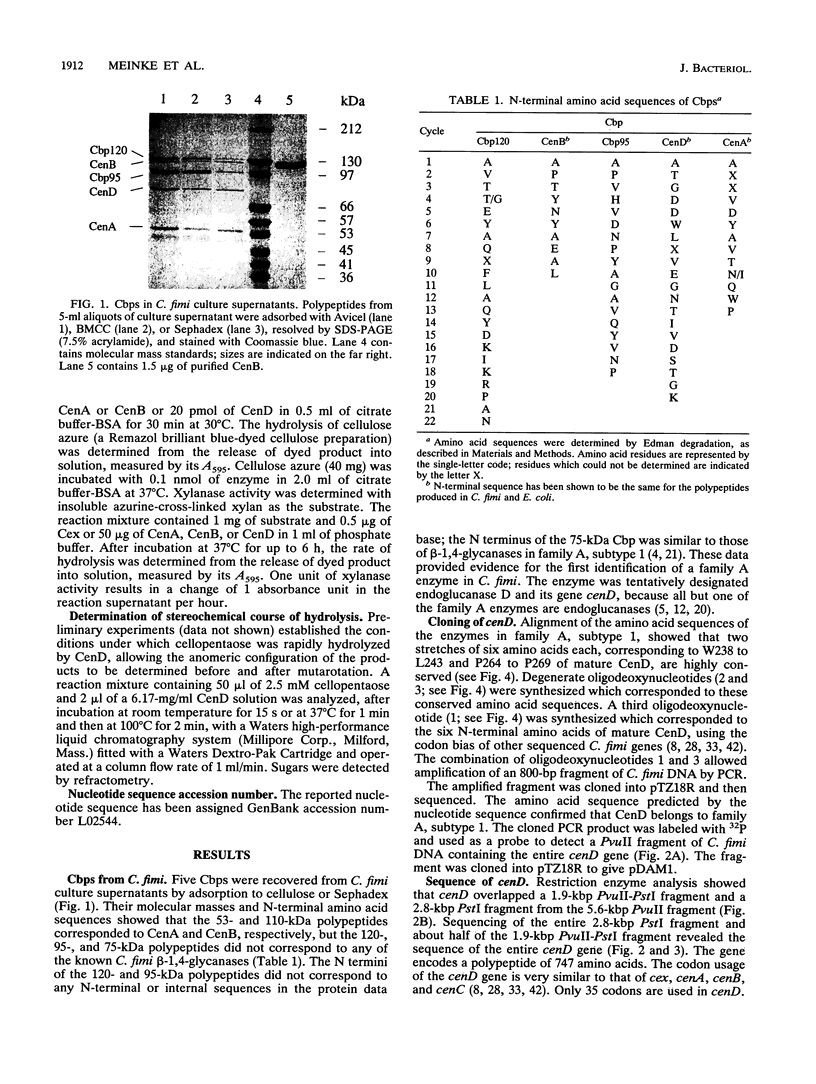
Five cellulose-binding polypeptides were detected in Cellulomonas fimi culture supernatants. Two of them are CenA and CenB, endo-beta-1,4-glucanases which have been characterized previously; the other three were previously uncharacterized polypeptides with apparent molecular masses of 120, 95, and 75 kDa. The 75-kDa cellulose-binding protein was designated endoglucanase D (CenD). The cenD gene was cloned and sequenced. It encodes a polypeptide of 747 amino acids. Mature CenD is 708 amino acids long and has a predicted molecular mass of 74,982 Da. Analysis of the predicted amino acid sequence of CenD shows that the enzyme comprises four domains which are separated by short linker polypeptides: an N-terminal catalytic domain of 405 amino acids, two repeated sequences of 95 amino acids each, and a C-terminal domain of 105 amino acids which is > 50% identical to the sequences of cellulose-binding domains in Cex, CenA, and CenB from C. fimi. Amino acid sequence comparison placed the catalytic domain of CenD in family A, subtype 1, of beta-1,4-glycanases. The repeated sequences are more than 40% identical to the sequences of three repeats in CenB and are related to the repeats of fibronectin type III. CenD hydrolyzed the beta-1,4-glucosidic bond with retention of anomeric configuration. The activities of CenD towards various cellulosic substrates were quite different from those of CenA and CenB.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baird S. D., Hefford M. A., Johnson D. A., Sung W. L., Yaguchi M., Seligy V. L. The Glu residue in the conserved Asn-Glu-Pro sequence of two highly divergent endo-beta-1,4-glucanases is essential for enzymatic activity. Biochem Biophys Res Commun. 1990 Jun 29;169(3):1035–1039. doi: 10.1016/0006-291x(90)91998-8. [DOI] [PubMed] [Google Scholar]
- Barras F., Bortoli-German I., Bauzan M., Rouvier J., Gey C., Heyraud A., Henrissat B. Stereochemistry of the hydrolysis reaction catalyzed by endoglucanase Z from Erwinia chrysanthemi. FEBS Lett. 1992 Mar 30;300(2):145–148. doi: 10.1016/0014-5793(92)80183-h. [DOI] [PubMed] [Google Scholar]
- Béguin P. Molecular biology of cellulose degradation. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- Coutinho J. B., Gilkes N. R., Warren R. A., Kilburn D. G., Miller R. C., Jr The binding of Cellulomonas fimi endoglucanase C (CenC) to cellulose and Sephadex is mediated by the N-terminal repeats. Mol Microbiol. 1992 May;6(9):1243–1252. doi: 10.1111/j.1365-2958.1992.tb01563.x. [DOI] [PubMed] [Google Scholar]
- Coutinho J. B., Moser B., Kilburn D. G., Warren R. A., Miller R. C., Jr Nucleotide sequence of the endoglucanase C gene (cenC) of Cellulomonas fimi, its high-level expression in Escherichia coli, and characterization of its products. Mol Microbiol. 1991 May;5(5):1221–1233. doi: 10.1111/j.1365-2958.1991.tb01896.x. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Claeyssens M., Aebersold R., Henrissat B., Meinke A., Morrison H. D., Kilburn D. G., Warren R. A., Miller R. C., Jr Structural and functional relationships in two families of beta-1,4-glycanases. Eur J Biochem. 1991 Dec 5;202(2):367–377. doi: 10.1111/j.1432-1033.1991.tb16384.x. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes N. R., Jervis E., Henrissat B., Tekant B., Miller R. C., Jr, Warren R. A., Kilburn D. G. The adsorption of a bacterial cellulase and its two isolated domains to crystalline cellulose. J Biol Chem. 1992 Apr 5;267(10):6743–6749. [PubMed] [Google Scholar]
- Gilkes N. R., Warren R. A., Miller R. C., Jr, Kilburn D. G. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J Biol Chem. 1988 Jul 25;263(21):10401–10407. [PubMed] [Google Scholar]
- Greenberg N. M., Warren R. A., Kilburn D. G., Miller R. C., Jr Regulation and initiation of cenB transcripts of Cellulomonas fimi. J Bacteriol. 1987 Oct;169(10):4674–4677. doi: 10.1128/jb.169.10.4674-4677.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg N. M., Warren R. A., Kilburn D. G., Miller R. C., Jr Regulation, initiation, and termination of the cenA and cex transcripts of Cellulomonas fimi. J Bacteriol. 1987 Feb;169(2):646–653. doi: 10.1128/jb.169.2.646-653.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Hazlewood G. P., Huskisson N. S., Durrant A. J., Gilbert H. J. Conserved serine-rich sequences in xylanase and cellulase from Pseudomonas fluorescens subspecies cellulosa: internal signal sequence and unusual protein processing. Mol Microbiol. 1989 Sep;3(9):1211–1219. doi: 10.1111/j.1365-2958.1989.tb00271.x. [DOI] [PubMed] [Google Scholar]
- Hansen C. K. Fibronectin type III-like sequences and a new domain type in prokaryotic depolymerases with insoluble substrates. FEBS Lett. 1992 Jun 29;305(2):91–96. doi: 10.1016/0014-5793(92)80871-d. [DOI] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991 Dec 1;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B., Claeyssens M., Tomme P., Lemesle L., Mornon J. P. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989 Sep 1;81(1):83–95. doi: 10.1016/0378-1119(89)90339-9. [DOI] [PubMed] [Google Scholar]
- Kellett L. E., Poole D. M., Ferreira L. M., Durrant A. J., Hazlewood G. P., Gilbert H. J. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990 Dec 1;272(2):369–376. doi: 10.1042/bj2720369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langsford M. L., Gilkes N. R., Singh B., Moser B., Miller R. C., Jr, Warren R. A., Kilburn D. G. Glycosylation of bacterial cellulases prevents proteolytic cleavage between functional domains. FEBS Lett. 1987 Dec 10;225(1-2):163–167. doi: 10.1016/0014-5793(87)81150-x. [DOI] [PubMed] [Google Scholar]
- Lever M. A new reaction for colorimetric determination of carbohydrates. Anal Biochem. 1972 May;47(1):273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- Meinke A., Braun C., Gilkes N. R., Kilburn D. G., Miller R. C., Jr, Warren R. A. Unusual sequence organization in CenB, an inverting endoglucanase from Cellulomonas fimi. J Bacteriol. 1991 Jan;173(1):308–314. doi: 10.1128/jb.173.1.308-314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke A., Gilkes N. R., Kilburn D. G., Miller R. C., Jr, Warren R. A. Bacterial cellulose-binding domain-like sequences in eucaryotic polypeptides. Protein Seq Data Anal. 1991 Dec;4(6):349–353. [PubMed] [Google Scholar]
- Meinke A., Gilkes N. R., Kilburn D. G., Miller R. C., Jr, Warren R. A. Multiple domains in endoglucanase B (CenB) from Cellulomonas fimi: functions and relatedness to domains in other polypeptides. J Bacteriol. 1991 Nov;173(22):7126–7135. doi: 10.1128/jb.173.22.7126-7135.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke A., Schmuck M., Gilkes N. R., Kilburn D. G., Miller R. C., Jr, Warren R. A. The tertiary structure of endo-beta-1,4-glucanase B (CenB), a multidomain cellulase from the bacterium Cellulomonas fimi. Glycobiology. 1992 Aug;2(4):321–326. doi: 10.1093/glycob/2.4.321. [DOI] [PubMed] [Google Scholar]
- Moser B., Gilkes N. R., Kilburn D. G., Warren R. A., Miller R. C., Jr Purification and characterization of endoglucanase C of Cellulomonas fimi, cloning of the gene, and analysis of in vivo transcripts of the gene. Appl Environ Microbiol. 1989 Oct;55(10):2480–2487. doi: 10.1128/aem.55.10.2480-2487.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill G., Goh S. H., Warren R. A., Kilburn D. G., Miller R. C., Jr Structure of the gene encoding the exoglucanase of Cellulomonas fimi. Gene. 1986;44(2-3):325–330. doi: 10.1016/0378-1119(86)90197-6. [DOI] [PubMed] [Google Scholar]
- Owolabi J. B., Beguin P., Kilburn D. G., Miller R. C., Warren R. A. Expression in Escherichia coli of the Cellulomonas fimi Structural Gene for Endoglucanase B. Appl Environ Microbiol. 1988 Feb;54(2):518–523. doi: 10.1128/aem.54.2.518-523.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem. 1974 May;59(1):277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Suzuki K., Oyanagi W., Ohnishi K., Tanaka H. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J Biol Chem. 1990 Sep 15;265(26):15659–15665. [PubMed] [Google Scholar]
- Whittle D. J., Kilburn D. G., Warren R. A., Miller R. C., Jr Molecular cloning of a Cellulomonas fimi cellulose gene in Escherichia coli. Gene. 1982 Feb;17(2):139–145. doi: 10.1016/0378-1119(82)90066-x. [DOI] [PubMed] [Google Scholar]
- Wong W. K., Gerhard B., Guo Z. M., Kilburn D. G., Warren A. J., Miller R. C., Jr Characterization and structure of an endoglucanase gene cenA of Cellulomonas fimi. Gene. 1986;44(2-3):315–324. doi: 10.1016/0378-1119(86)90196-4. [DOI] [PubMed] [Google Scholar]