Abstract
In Rhizobium meliloti, an aspartate aminotransferase (AspAT) encoded within a 7.3-kb HindIII fragment was previously shown to be required for symbiotic nitrogen fixation and aspartate catabolism (V. K. Rastogi and R.J. Watson, J. Bacteriol. 173:2879-2887, 1991). A gene coding for an aromatic aminotransferase located within an 11-kb HindIII fragment was found to complement the AspAT deficiency when overexpressed. The genes encoding these two aminotransferases, designated aatA and tatA, respectively, have been localized by subcloning and transposon Tn5 mutagenesis. Sequencing of the tatA gene revealed that it encodes a protein homologous to an Escherichia coli aromatic aminotransferase and most of the known AspAT enzymes. However, sequencing of the aatA gene region revealed two overlapping open reading frames, neither of which encoded an enzyme with homology to the typical AspATs. Polymerase chain reaction was used to selectively generate one of the candidate sequences for subcloning. The cloned fragment complemented the original nitrogen fixation and aspartate catabolism defects and was shown to encode an AspAT with the expected properties. Sequence analysis showed that the aatA protein has homology to AspATs from two thermophilic bacteria and the eukaryotic tyrosine aminotransferases. These aminotransferases form a distinct class in which only 13 amino acids are conserved in comparison with the well-known AspAT family. DNA homologous to the aatA gene was found to be present in Agrobacterium tumefaciens and other rhizobia but not in Klebsiella pneumoniae or E. coli.
Full text
PDF



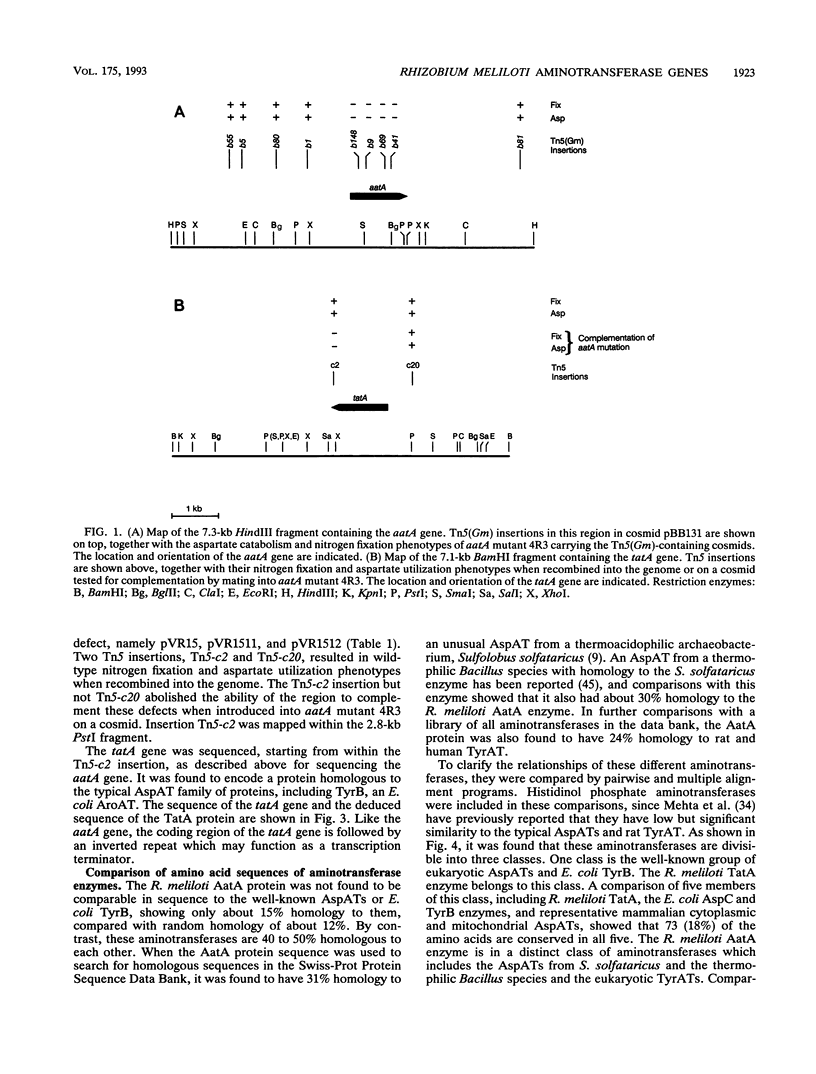





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appels M. A., Haaker H. Glutamate Oxaloacetate Transaminase in Pea Root Nodules : Participation in a Malate/Aspartate Shuttle between Plant and Bacteroid. Plant Physiol. 1991 Mar;95(3):740–747. doi: 10.1104/pp.95.3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Birolo L., Arnone M. I., Cubellis M. V., Andreotti G., Nitti G., Marino G., Sannia G. The active site of Sulfolobus solfataricus aspartate aminotransferase. Biochim Biophys Acta. 1991 Nov 15;1080(3):198–204. doi: 10.1016/0167-4838(91)90002-h. [DOI] [PubMed] [Google Scholar]
- Cronin V. B., Maras B., Barra D., Doonan S. The amino acid sequence of the aspartate aminotransferase from baker's yeast (Saccharomyces cerevisiae). Biochem J. 1991 Jul 15;277(Pt 2):335–340. doi: 10.1042/bj2770335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubellis M. V., Rozzo C., Nitti G., Arnone M. I., Marino G., Sannia G. Cloning and sequencing of the gene coding for aspartate aminotransferase from the thermoacidophilic archaebacterium Sulfolobus solfataricus. Eur J Biochem. 1989 Dec 8;186(1-2):375–381. doi: 10.1111/j.1432-1033.1989.tb15219.x. [DOI] [PubMed] [Google Scholar]
- Dietrich J. B. Tyrosine aminotransferase: a transaminase among others? Cell Mol Biol. 1992 Apr;38(2):95–114. [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff T. T., Long S. R. Rhizobium meliloti nodulation genes: identification of nodDABC gene products, purification of nodA protein, and expression of nodA in Rhizobium meliloti. J Bacteriol. 1985 Nov;164(2):591–599. doi: 10.1128/jb.164.2.591-599.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan T. M., Hartweig E., LeMieux K., Bergman K., Walker G. C., Signer E. R. General transduction in Rhizobium meliloti. J Bacteriol. 1984 Jul;159(1):120–124. doi: 10.1128/jb.159.1.120-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan T. M., Kunkel B., De Vos G. F., Signer E. R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986 Jul;167(1):66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford G. C., Eichele G., Jansonius J. N. Three-dimensional structure of a pyridoxal-phosphate-dependent enzyme, mitochondrial aspartate aminotransferase. Proc Natl Acad Sci U S A. 1980 May;77(5):2559–2563. doi: 10.1073/pnas.77.5.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotheringham I. G., Dacey S. A., Taylor P. P., Smith T. J., Hunter M. G., Finlay M. E., Primrose S. B., Parker D. M., Edwards R. M. The cloning and sequence analysis of the aspC and tyrB genes from Escherichia coli K12. Comparison of the primary structures of the aspartate aminotransferase and aromatic aminotransferase of E. coli with those of the pig aspartate aminotransferase isoenzymes. Biochem J. 1986 Mar 15;234(3):593–604. doi: 10.1042/bj2340593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantt J. S., Larson R. J., Farnham M. W., Pathirana S. M., Miller S. S., Vance C. P. Aspartate aminotransferase in effective and ineffective alfalfa nodules : cloning of a cDNA and determination of enzyme activity, protein, and mRNA levels. Plant Physiol. 1992 Mar;98(3):868–878. doi: 10.1104/pp.98.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand D. H., Steinberg R. A. Escherichia coli mutants deficient in the aspartate and aromatic amino acid aminotransferases. J Bacteriol. 1977 Apr;130(1):429–440. doi: 10.1128/jb.130.1.429-440.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf-Hausner U., Wilson K. J., Christen P. The covalent structure of mitochondrial aspartate aminotransferase from chicken. Identification of segments of the polypeptide chain invariant specifically in the mitochondrial isoenzyme. J Biol Chem. 1983 Jul 25;258(14):8813–8826. [PubMed] [Google Scholar]
- Griffith S. M., Vance C. P. Aspartate aminotransferase in alfalfa root nodules : I. Purification and partial characterization. Plant Physiol. 1989 Aug;90(4):1622–1629. doi: 10.1104/pp.90.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harutyunyan E. G., Malashkevich V. N., Tersyan S. S., Kochkina V. M., Torchinsky YuM, Braunstein A. E. Three-dimensional structure at 3.2 A resolution of the complex of cytosolic aspartate aminotransferase from chicken heart with 2-oxoglutarate. FEBS Lett. 1982 Feb 8;138(1):113–116. doi: 10.1016/0014-5793(82)80407-9. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hynes M. F., Simon R., Pühler A. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtC58. Plasmid. 1985 Mar;13(2):99–105. doi: 10.1016/0147-619x(85)90062-9. [DOI] [PubMed] [Google Scholar]
- Jensen R. A., Calhoun D. H. Intracellular roles of microbial aminotransferases: overlap enzymes across different biochemical pathways. Crit Rev Microbiol. 1981;8(3):229–266. doi: 10.3109/10408418109085080. [DOI] [PubMed] [Google Scholar]
- Kirsch J. F., Eichele G., Ford G. C., Vincent M. G., Jansonius J. N., Gehring H., Christen P. Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. J Mol Biol. 1984 Apr 15;174(3):497–525. doi: 10.1016/0022-2836(84)90333-4. [DOI] [PubMed] [Google Scholar]
- Kittell B. L., Helinski D. R., Ditta G. S. Aromatic aminotransferase activity and indoleacetic acid production in Rhizobium meliloti. J Bacteriol. 1989 Oct;171(10):5458–5466. doi: 10.1128/jb.171.10.5458-5466.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Marino G., Nitti G., Arnone M. I., Sannia G., Gambacorta A., De Rosa M. Purification and characterization of aspartate aminotransferase from the thermoacidophilic archaebacterium Sulfolobus solfataricus. J Biol Chem. 1988 Sep 5;263(25):12305–12309. [PubMed] [Google Scholar]
- Mattes U., Jaussi R., Ziak M., Juretic N., Lindenmann J. M., Christen P. Structure of cDNA of cytosolic aspartate aminotransferase of chicken and its expression in E. coli. Biochimie. 1989 Apr;71(4):411–416. doi: 10.1016/0300-9084(89)90171-5. [DOI] [PubMed] [Google Scholar]
- Mehta P. K., Hale T. I., Christen P. Evolutionary relationships among aminotransferases. Tyrosine aminotransferase, histidinol-phosphate aminotransferase, and aspartate aminotransferase are homologous proteins. Eur J Biochem. 1989 Dec 8;186(1-2):249–253. doi: 10.1111/j.1432-1033.1989.tb15202.x. [DOI] [PubMed] [Google Scholar]
- Myers E. W., Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988 Mar;4(1):11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Rastogi V. K., Watson R. J. Aspartate aminotransferase activity is required for aspartate catabolism and symbiotic nitrogen fixation in Rhizobium meliloti. J Bacteriol. 1991 May;173(9):2879–2887. doi: 10.1128/jb.173.9.2879-2887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakawa C., Yoshikawa M. A series of Tn5 variants with various drug-resistance markers and suicide vector for transposon mutagenesis. Gene. 1987;56(2-3):283–288. doi: 10.1016/0378-1119(87)90145-4. [DOI] [PubMed] [Google Scholar]
- Stowers M. D. Carbon metabolism in Rhizobium species. Annu Rev Microbiol. 1985;39:89–108. doi: 10.1146/annurev.mi.39.100185.000513. [DOI] [PubMed] [Google Scholar]
- Sung M. H., Tanizawa K., Tanaka H., Kuramitsu S., Kagamiyama H., Hirotsu K., Okamoto A., Higuchi T., Soda K. Thermostable aspartate aminotransferase from a thermophilic Bacillus species. Gene cloning, sequence determination, and preliminary x-ray characterization. J Biol Chem. 1991 Feb 5;266(4):2567–2572. [PubMed] [Google Scholar]
- Ta T. C., Faris M. A., Macdowall F. D. Pathways of Nitrogen Metabolism in Nodules of Alfalfa (Medicago sativa L.). Plant Physiol. 1986 Apr;80(4):1002–1005. doi: 10.1104/pp.80.4.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M. K., Kahn M. L. Isolation and analysis of a cDNA clone that encodes an alfalfa (Medicago sativa) aspartate aminotransferase. Mol Gen Genet. 1991 Dec;231(1):97–105. doi: 10.1007/BF00293827. [DOI] [PubMed] [Google Scholar]
- Watson R. J. Analysis of the C4-dicarboxylate transport genes of Rhizobium meliloti: nucleotide sequence and deduced products of dctA, dctB, and dctD. Mol Plant Microbe Interact. 1990 May-Jun;3(3):174–181. doi: 10.1094/mpmi-3-174. [DOI] [PubMed] [Google Scholar]
- Watson R. J., Chan Y. K., Wheatcroft R., Yang A. F., Han S. H. Rhizobium meliloti genes required for C4-dicarboxylate transport and symbiotic nitrogen fixation are located on a megaplasmid. J Bacteriol. 1988 Feb;170(2):927–934. doi: 10.1128/jb.170.2.927-934.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]