Abstract
Several cDNAs isolated from brains of diapausing pupae of the flesh fly, Sarcophaga crassipalpis, show expression patterns unique to diapause. To isolate such cDNAs a diapause pupal brain cDNA library was screened by using an elimination hybridization technique, and cDNAs that did not hybridize with cDNA probes constructed from the RNA of nondiapausing pupae were selected for further screening. The 95 clones that did not hybridize in the initial library screen were selected for further characterization. These clones were then screened against diapause and nondiapause pupal poly(A)+ Northern blots. The secondary screen identified 4 diapause-up-regulated clones, 7 diapause-down-regulated clones, 8 clones expressed equally in both diapause and nondiapause, and 75 clones without detectable expression. The diapause-up-regulated and down-regulated clones were further characterized by partial DNA sequencing and identity searches by using GenBank. Identities between our cloned cDNAs and other genes included those linked to cell cycle progression, stress responses, and DNA repair processes. The results suggest that insect diapause is not merely a shutdown of gene expression but is a unique, developmental pathway characterized by the expression of a novel set of genes.
Keywords: insect, cell cycle, proliferating cell nuclear antigen, protein kinase, heat shock proteins
Most insect species have evolved a period of developmental arrest (diapause) that enables them to circumvent seasonal periods of adversity. For insects in the temperate zone, winter is the season most consistently avoided. It is not at all uncommon for a species to spend 9–10 months in diapause, with only 2–3 months of summer devoted to active development and reproduction (1, 2). Within a single species the potential for diapause usually is restricted to a specific developmental stage, but embryonic, larval, pupal, and adult diapauses are all well documented. Whether the insect will enter diapause usually is dictated by the day-length perceived by the insect at an earlier stage of development. Long day-lengths frequently channel the insect toward uninterrupted development during late spring and early summer, whereas the short day-lengths of late summer and autumn program the entry into diapause. Day-length thus presides over the hormonal mechanisms that direct the insect toward either diapause or nondiapause development (3).
Like hibernating mammals, insects in diapause are totally dependent on the energy reserves that have been sequestered during earlier active phases of the life cycle. Suppression of metabolism enables the insect to stretch its food reserves to bridge the unfavorable period. Survival during diapause may also be enhanced by the synthesis of polyols and other cryoprotective agents that reduce injury at low temperatures. Thus, the diapause and nondiapause phases of the insect’s life cycle represent striking contrasts. What these differences may mean at the molecular level remains largely unknown. Whether diapause is simply a shutdown in gene expression or whether it represents a unique pattern of gene expression has long been debated. We addressed this question in the flesh fly, Sarcophaga crassipalpis, by searching for differences in gene expression in the brains of diapausing and nondiapausing pupae.
Our search focused on the brain because it is this tissue that is responsible both for receiving the environmental cues involved in inducing diapause as well as for executing the diapause program (3, 4). The flesh flies used in this study enter an overwintering pupal diapause in response to cues of short day-length received during late embryonic and early larval life (5–7). Hormonally, this diapause can be characterized as an ecdysteroid deficiency; the brain fails to stimulate the prothoracic gland to synthesize ecdysteroids and hence development is halted until the synthesis of ecdysteroids again is invoked at the end of diapause (8–10). An examination of brain proteins synthesized during diapause (11) demonstrated that diapause represents both a partial shutdown in gene expression (far fewer proteins were expressed in brains of diapausing pupae than in brains of nondiapausing pupae) and the expression of a unique set of genes (a cluster of 14 proteins was expressed only in brains of diapausing pupae). In this study, we isolated diapause-specific genes and evaluated the abundance of such genes among the total pool of mRNA expressed in the brain during diapause.
MATERIALS AND METHODS
Colony Maintenance.
The colony of S. crassipalpis was maintained as described (12). The decision to enter pupal diapause is made during a 4-day window in late embryonic and early larval development (6). To produce nondiapause pupae, adults were maintained at a 15-hr light/9-hr dark (15L:9D) cycle and 25°C. After larviposition, the larvae were reared at the same conditions as the adults for the first 4 days and were then transferred to 12L:12D at 20°C for the remainder of development. To produce diapause pupae, adults were maintained at 12L:12D at 25°C. After larviposition, the larvae were transferred immediately to 12L:12D at 20°C.
RNA Purification.
Pupal brains were homogenized in TRIzol reagent (GIBCO/BRL) and stored at −70°C. RNA was purified by using the standard TRIzol protocol (GIBCO/BRL). The total brain RNA was resuspended in diethyl-pyrocarbonate-treated water. Total body RNA was dissolved in binding buffer [10 mM Tris⋅HCl, pH 7.5/1 mM EDTA/0.3 M NaCl/0.1% (wt/vol) SDS] and passed through a column of oligo(dT) cellulose. The column was rinsed twice with binding buffer and then eluted with elution buffer [10 mM Tris⋅HCl, pH 7.5/1 mM EDTA/0.1% (wt/vol) SDS]. The poly(A)+ RNA was quantitated, precipitated, and resuspended in formamide at 2 μg/μl.
Library Construction and Isolation of cDNAs.
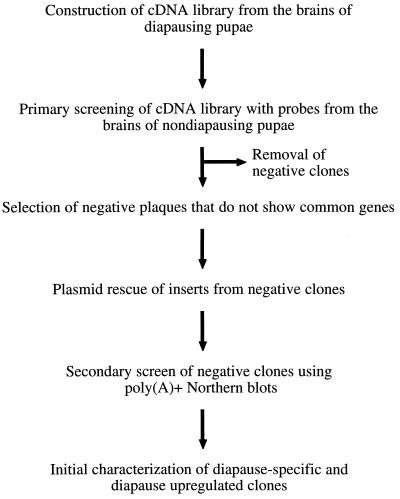
The overall strategy for the isolation of diapause-specific genes from the diapause cDNA library is shown in Fig. 1. The commercial library was prepared by isolating poly(A)+ RNA from 100 diapausing pupal brains using an oligo(dT) primer for first-strand cDNA synthesis (CLONTECH). A titer of 1.8 × 106 pfu/ml was obtained with 1.75 × 106 (97%) as independent recombinant clones as determined by 5-bromo-4-chloro-3-indolyl β-d-galactoside screening. The primary library screening was performed by making a complex mixture of probes from total brain RNA of nondiapause-destined pupae. Probes were constructed by performing first-strand synthesis from total RNA using oligo(dT) primers and a mixture of biotin-labeled dNTPs (Stratagene). Library screening was performed according to the manufacturer’s instructions (CLONTECH). Phage that did not show hybridization were then picked as putative diapause-specific clones. The phage were subjected to an in vivo excision process that resulted in the cloning of the cDNA.
Figure 1.
Schematic diagram of the elimination hybridization technique used to isolate putative diapause-specific clones.
Northern Blotting.
Equivalent amounts of poly(A)+ RNA from diapausing and nondiapausing whole pupae were loaded on a 1.2% formaldehyde (0.41 M) denaturing gel and transferred to a nylon membrane by using a Schleicher & Schuell Turboblotter via downward capillary action. Prehybridization and hybridization were carried out in a 1× hybridization buffer [0.5 M NaCl/0.1 M NaPO4, pH 7.0/6 mM EDTA/1% (wt/vol) SDS] at 65°C. cDNA probes were constructed by using the Rad Prime system (GIBCO/BRL) in the presence of [32P]dCTP (3,000 Ci/mmol; 1 Ci = 37 GBq). Washes consisted of a single wash in 1/4× hybridization buffer at 65°C and two washes in 1/4× hybridization buffer without SDS at 65°C. The membranes were then wrapped in Saran wrap and exposed to Kodak XAR5 film at −70°C. To ensure that equivalent RNA was loaded in each lane, samples were quantified spectrophotometrically; in addition, ethidium bromide was added to all samples and a photograph was taken to compare rRNA band intensity.
DNA Sequencing.
DNA sequencing was performed on all diapause-up-regulated and down-regulated genes, as well as those showing equal signals in both diapause and nondiapause brains. The DNA was sequenced at the University of Georgia at Athens on an Applied Biosystems 373A DNA sequencer by using dye terminator chemistry according to the manufacturer’s standard protocol. The blast search program was used to search the GenBank sequence repository for sequence identities.
RESULTS
Identification and Isolation of Putative Diapause-Specific cDNAs.
Initial screening of 450 plaques yielded 355 (79%) that hybridized strongly to nondiapause pupal brain cDNAs and 95 (21%) that failed to hybridize to the RNA blots. These 95 clones were selected as putative diapause-specific clones, and each was given a number, plasmid Sarcophaga crassipalpis diapause 1 (pScD1) to pScD95.
Secondary Screen Using Northern Blotting.
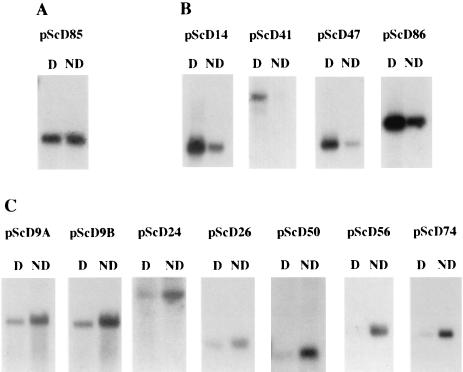
Northern blotting of total pupal poly(A)+ RNA from diapause and nondiapause pupae was used to determine whether any of the 95 clones were associated exclusively with diapause. The clones were then grouped according to four hybridization patterns: (i) up-regulation in diapause, (ii) down-regulation in diapause, (iii) hybridization equally in diapause and nondiapause, and (iv) no hybridization to diapause or nondiapause. Four clones (4.2%)—pScD14, pScD41, pScD47, and pScD86—were diapause-up-regulated (Fig. 2B). In each of these cases, some hybridization also was seen to the nondiapause RNA. With a longer exposure, even pScD41 showed a weak signal in the nondiapause lane. Seven clones (7.4%)—pScD9A, pScD9B, pScD24, pScD26, pScD50, pScD56, and pScD74—produced a weaker signal to diapause RNA than to nondiapause RNA and were classified as diapause-down-regulated (Fig. 2C). Eight clones (8.4%)—pScD20, pScD21, pScD22, pScD45A, pScD45B, pScD73, pScD85, and pScD93—produced a signal of equal intensity in both the diapause and nondiapause lanes. pScD85, shown in Fig. 2A, is a representative of this group. The final group consisted of the 75 pScD clones (80.0%) that did not produce a signal in either the diapause or nondiapause lanes (data not shown). Transcript sizes of the pScD clones mentioned are listed in Table 1.
Figure 2.
Hybridization of cDNA probes to Northern blots. Four micrograms of diapause (D) and nondiapause (ND) pupal poly(A)+ RNA were hybridized with 32P-labeled probes from the indicated clones. (A) pScD85 is a representative of clones that hybridized equally to diapause and nondiapause pupal RNA. (B) pScD14, pScD41, pScD47, and pScD86 clones are diapause-up-regulated. (C) pScD9A, pScD9B, pScD24, pScD26, pScD50, pScD56, and pScD74 clones are diapause-down-regulated. Equal loading of RNA was ensured by spectrophotometric analysis and ethidium bromide staining of the gel.
Table 1.
Sequence identities and transcript sizes of diapause-up-regulated, diapause-down-regulated clones and clones that are expressed equally in diapause and nondiapause individuals
| Clone | Transcript size, kb | Protein | Identity, % |
|---|---|---|---|
| Diapause-up-regulated | |||
| pScD14 | 1.4 | Small heat shock protein 23 | 85 |
| pScD41 | 2.4 | No identity | |
| pScD47 | 1.4 | No identity | |
| pScD86 | 2.1 | Apurinic-apyrimidinic endonuclease* | 88 |
| Diapause-down-regulated | |||
| pScD9A | 1.6 | Elastin-like protein† | 90 |
| pScD9B | 1.5 | Elastin-like protein† | 63 |
| pScD24 | 2.0 | No identity | |
| pScD26 | 1.0 | No identity | |
| pScD50 | 0.7 | No identity | |
| pScD56 | 1.3 | Proliferating cell nuclear antigen | 82 |
| pScD74 | 1.2 | Protein kinase (GPK2) | 73 |
| Expressed equally in diapause and nondiapause | |||
| pScD20 | 1.3 | Apurinic-apyrimidinic endonuclease* | 90 |
| pScD21 | 1.0 | Apurinic-apyrimidinic endonuclease* | 90 |
| pScD22 | 1.6 | Not sequenced | |
| pScD45A | 0.7 | Not sequenced | |
| pScD45B | 1.8 | Not sequenced | |
| pScD73 | 1.0 | Not sequenced | |
| pScD85 | 1.5 | Hsp 70 cognate | 93 |
| pScD93 | 1.4 | No identity |
The % identities are based on partial DNA sequences (600–800 bp per clone). Scores are based on the default settings for blast parameters of % amino acid identity. All identities are to Drosophila melanogaster, except pScD74, which is to Dictyostelium discoideum.
Clones have high identity to each other after partial sequence analysis, but restriction mapping revealed that these are distinct clones.
Clones have high identity to the same sequence, but pScD9A contains an internal insertion of 70 bp not contained in pScD9B.
Sequence Identity of cDNAs.
Partial sequence data were obtained for all of the diapause-up-regulated and diapause-down-regulated clones, and percentage of identities to sequences deposited in GenBank are listed in Table 1. Diapause-up-regulated clones pScD14 and pScD86 showed high identity to a Drosophila melanogaster small heat shock protein (13) and a D. melanogaster apurinic-apyrimidinic endonuclease (14), respectively. The other two diapause-up-regulated clones showed no identity to known sequences.
The diapause-down-regulated clone, pScD56, has a high level of identity to proliferating cell nuclear antigen (PCNA) from D. melanogaster (15). pScD9A and pScD9B appear to be the same clone; however, pScD9A contains a 70-nt stretch that is not present in pScD9B. Both of the pScD9 sequences have high identity to an elastin-like protein from D. melanogaster (16). The similarity of the two clones raises the possibility that transcripts are alternatively spliced. pScD74 has high identity to a protein kinase from the slime mold Dictyostelium discoideum. The remaining diapause-down-regulated clones showed no identity to known sequences.
We also have partially sequenced pScD20, pScD21, pScD85, and pScD93, all of which hybridized equally to diapause and nondiapause RNA. pScD20 and pScD21 have high identity to an apurinic-apyrimidinic endonuclease (14), whereas pScD85 has high identity to the heat shock 70 cognate (17).
DISCUSSION
The evidence we present here, combined with previous work on brain proteins (11), suggests that diapause should be regarded as a unique developmental pathway rather than a simple shutdown of gene expression. Although certain genes clearly are down-regulated during diapause, others are strongly up-regulated. From the sample of 95 clones initially isolated, 4.2% proved to be diapause-up-regulated. This percentage is remarkably similar to the incidence of diapause-specific brain proteins (9%) previously reported for S. crassipalpis based on two-dimensional electrophoresis of 35S-pulse-labeled proteins (11). A similar estimate of diapause-specific gene activity in this species was obtained by differential display of mRNA (ref. 18; K.H.J. and D.L.D., unpublished observations): a sample of 332 well defined transcripts from diapause brains included 35 (10.5%) that appeared to be diapause-specific. That this technique generates many false positives suggests that the 10.5% incidence is likely to be a high estimate. Yet, all three techniques (library screening and Northern blot analysis, two-dimensional electrophoresis of brain proteins, and differential display) indicate the presence of a discrete set of genes that are expressed in association with insect diapause. Our estimate suggests that diapause-specific and/or diapause-up-regulated genes represent 4–10% of the genes being expressed during diapause in the pupal brains of the flesh fly.
Several previous studies have documented the presence of diapause-associated proteins from the fat body and hemolymph of diapausing insects (19–24). These all have proven to be storage proteins that are synthesized before the onset of diapause and are then utilized when development resumes at the termination of diapause (25). Also, a unique midgut protein is synthesized during diapause in the gypsy moth, Lymantria dispar (26). Although all such proteins are potentially important for maintenance of diapause, their site of synthesis, the fat body or midgut, suggests they represent events that are downstream from the site of diapause regulation. As the center for diapause regulation, the brain is the most likely site in which to seek regulatory genes, yet we can assume that many diapause-specific genes expressed in the brain also will prove to be involved in coordinating downstream events including the maintenance of diapause.
Only two of the four diapause-up-regulated clones we have isolated show high identity to known genes. pScD14, the clone that shows high identity (85%) to a small heat shock protein, could be involved in the cell cycle arrest that occurs during diapause (27). Yeast and human β lymphocytes express certain small heat shock proteins during cell cycle arrest (28–30). Cold shock also elicits the expression of small heat shock proteins in S. crassipalpis (31) and in the gypsy moth L. dispar (32). At the onset of diapause in S. crassipalpis, cold tolerance increases dramatically (33), a response that is consistent with the expression of the pScD14 transcript we observed in this study. Subsequently, full-length cloning and sequencing of pScD14 have been accomplished (GenBank accession no. U96099), and this gene is consistently up-regulated during the entire course of diapause. pScD86, the second diapause-up-regulated clone with a high identity with a known gene, appears to be an apurinic-apyrimidinic endonuclease. This enzyme is critical to DNA-repair processes and acts at apurinic-apyrimidinic sites on DNA to initiate nucleotide excision repair (14). Possibly such repair is critical for an insect that remains exposed to harsh environmental conditions for such a prolonged duration.
The identity of diapause-down-regulated clones is of equal importance. The identity of pScD56 as PCNA is of particular interest because of the role of PCNA in regulating development and cell cycle status (34). During diapause, the cells of the flesh fly brain are in a G0/G1 cell cycle arrest (27). Full-length cloning and sequencing of PCNA have been accomplished (GenBank accession no. AF020427), and its expression during diapause currently is under investigation. The down-regulation of PCNA during diapause and the rapid onset of its expression at diapause termination, when the cells again begin to cycle, suggest a potential role for PCNA in regulating the cell cycle arrest associated with diapause. The identity of another diapause-down-regulated clone, pScD74, also suggests a potentially important role. This clone shows high sequence identity to protein kinases, an enzyme family well known to play critical roles in the regulation of development.
The physiological roles and identities for most of the up-regulated and down-regulated clones remain to be defined, and many additional clones of interest are yet to be isolated and characterized. Transcripts from many clones within the brain are likely to be expressed at very low levels that would not have been detected with our procedures. Of the 95 clones isolated, 75 did not detect homologous RNA sequences in either diapause or nondiapause individuals when used as probes.
The isolation of diapause-regulated clones should enable us to assess the features of diapause regulation in flesh flies that are shared with other organisms having diapause stages in their life cycles. Diapause is widespread in insects and, depending on the species, may occur in embryonic stages, larvae, pupae, or adults. Diapause-like states also are common in other arthropods and in a wide range of invertebrates. Perhaps the best known diapause-like state in other invertebrates is the dauer larval stage of the nematode, Caenorhabditis elegans, a nonfeeding, nongrowing larval stage that is initiated in response to starvation and crowding. The use of mutants of the dauer formation (daf) genes has allowed investigators to order these genes in a developmental hierarchy (35, 36). Of special importance is daf-2, a member of the insulin receptor family, that dramatically extends longevity (37, 38). From the work on C. elegans it is apparent that the dauer state represents a unique developmental pathway (39, 40), a conclusion that now also appears to be appropriate for insect diapause. It is not yet clear whether the diverse manifestations of diapause and related forms of dormancy share a common regulatory basis or have attained a similar developmental stasis by alternative regulatory mechanisms.
Acknowledgments
This research was supported in part by U.S. Department of Agriculture-National Research Initiative Grant No. 94–37302-0502
ABBREVIATION
- PCNA
proliferating cell nuclear antigen
Footnotes
References
- 1.Saunders D S. Insect Clocks. Oxford: Pergamon; 1982. [Google Scholar]
- 2.Tauber M J, Tauber C A, Masaki S. Seasonal Adaptations of Insects. New York: Oxford Univ. Press; 1986. [Google Scholar]
- 3.Denlinger D L. In: Comprehensive Insect Physiology, Biochemistry and Pharmacology. Kerkut G A, Gilbert L I, editors. Vol. 8. New York: Pergamon; 1985. pp. 353–412. [Google Scholar]
- 4.Giebultowicz J M, Denlinger D L. J Insect Physiol. 1986;32:161–166. [Google Scholar]
- 5.Saunders D S. J Insect Physiol. 1971;17:801–812. [Google Scholar]
- 6.Denlinger D L. J Insect Physiol. 1971;17:1815–1822. doi: 10.1016/0022-1910(71)90126-0. [DOI] [PubMed] [Google Scholar]
- 7.Gnagey A L, Denlinger D L. J Comp Physiol. 1984;154:91–96. [Google Scholar]
- 8.Fraenkel G, Hsiao C. J Insect Physiol. 1968;14:707–718. [Google Scholar]
- 9.Zdarek J, Denlinger D L. J Insect Physiol. 1975;21:1193–1202. doi: 10.1016/0022-1910(75)90087-6. [DOI] [PubMed] [Google Scholar]
- 10.Walker G P, Denlinger D L. J Insect Physiol. 1980;26:661–664. [Google Scholar]
- 11.Joplin K H, Yocum G D, Denlinger D L. J Insect Physiol. 1990;36:775–783. [Google Scholar]
- 12.Denlinger D L. Biol Bull. 1972;142:11–24. [Google Scholar]
- 13.Ingolia T D, Craig E A. Proc Natl Acad Sci USA. 1982;79:2360–2364. doi: 10.1073/pnas.79.7.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley M R, Venugopal S, Harless J, Deutsh W A. Mol Cell Biol. 1989;9:965–973. doi: 10.1128/mcb.9.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng L, Prelich G, Anderson C W, Stillman B, Fisher P A. J Biol Chem. 1990;265:11948–11954. [PubMed] [Google Scholar]
- 16.Li W, Filippov V A, Filippova M A, Jindra M, Sehnal F. Eur J Entomol. 1995;92:151–155. [Google Scholar]
- 17.Perkins L A, Doctor J S, Zhang K, Stinson L, Perrimon N, Craig E A. Mol Cell Biol. 1990;10:3232–3238. doi: 10.1128/mcb.10.6.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denlinger D L, Joplin K H, Flannagan R D, Tammariello S P, Zhang M,-L, Yocum G D, Lee K-Y. In: Molecular Mechanisms of Insect Metamorphosis and Diapause. Suzuki A, Kataoka H, Matsumoto S, editors. Tokyo: Industrial Publishing and Consulting Inc.; 1995. pp. 289–297. [Google Scholar]
- 19.Brown J J, Chippendale G M. Insect Biochem. 1978;8:359–367. [Google Scholar]
- 20.Brown J J. J Insect Physiol. 1980;26:487–495. [Google Scholar]
- 21.Peferoen M, Stynen D, De Loof A. Comp Biochem Physiol. 1982;72B:345–351. [Google Scholar]
- 22.Osir E O, Labongo L V, Unnithan G C. Arch Insect Biochem. 1989;11:173–187. [Google Scholar]
- 23.Salama M S, Miller T A. Arch Insect Biochem Physiol. 1992;21:1–11. doi: 10.1002/arch.940210102. [DOI] [PubMed] [Google Scholar]
- 24.Palli, S. R., Ladd, T. R., Ricci, A. R., Primavera, M., Mungrue, I. N., Pang, A. S. D. & Retnakaran, A. (1998) J. Insect Physiol., in press. [DOI] [PubMed]
- 25.Levenbook L. In: Comprehensive Insect Physiology, Biochemistry and Pharmacology. Kerkut G A, Gilbert L I, editors. Vol. 10. New York: Pergamon; 1985. pp. 307–346. [Google Scholar]
- 26.Lee K-Y, Denlinger D L. J Insect Physiol. 1996;42:423–431. [Google Scholar]
- 27.Tammariello, S. P. & Denlinger, D. L. (1998) Insect Biochem. Mol. Biol., in press. [DOI] [PubMed]
- 28.Kurtz S, Rossi J, Petko L, Lindquist S. Science. 1986;231:1154–1157. doi: 10.1126/science.3511530. [DOI] [PubMed] [Google Scholar]
- 29.Rossi J M, Lindquist S. J Cell Biol. 1989;108:425–439. doi: 10.1083/jcb.108.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spector N L, Samson W, Ryan C, Gribben J, Urba W, Welch W J, Nadler L M. J Immunol. 1992;148:1668–1673. [PubMed] [Google Scholar]
- 31.Joplin K H, Yocum G D, Denlinger D L. J Insect Physiol. 1990;36:825–834. [Google Scholar]
- 32.Yocum G D, Joplin K H, Denlinger D L. Arch Insect Biochem Physiol. 1991;18:239–249. [Google Scholar]
- 33.Lee R E, Jr, Denlinger D L. Physiol Entomol. 1985;10:309–315. [Google Scholar]
- 34.Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtsuka E, Tsurimoto T. J Biol Chem. 1995;270:22527–22534. doi: 10.1074/jbc.270.38.22527. [DOI] [PubMed] [Google Scholar]
- 35.Georgi L L, Albert P S, Riddle D L. Cell. 1990;61:635–645. doi: 10.1016/0092-8674(90)90475-t. [DOI] [PubMed] [Google Scholar]
- 36.Estevez M, Attisano L, Wrana J L, Albert P S, Massague J, Riddle D L. Nature (London) 1993;365:644–649. doi: 10.1038/365644a0. [DOI] [PubMed] [Google Scholar]
- 37.Wood W B, Johnson T E. Curr Biol. 1994;4:151–153. doi: 10.1016/s0960-9822(94)00036-9. [DOI] [PubMed] [Google Scholar]
- 38.Kimura K D, Tissenbaum H A, Liu Y, Ruvkun G. Science. 1997;277:942–945. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 39.Riddle D L, Swanson M M, Albert P S. Nature (London) 1981;290:668–672. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- 40.Gottlieb S, Ruvkun G. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]