Abstract
The Wnt-inducible homeobox gene Siamois is expressed in Xenopus embryos before gastrulation and is necessary for formation of the Spemann organizer. Here we show that 5′-flanking sequences of the Siamois coding region can specifically activate a heterologous reporter gene in dorsovegetal cells, thus mimicking Siamois’s endogenous expression. A 245-bp DNA fragment is sufficient for activation by both Wnts and endogenous inducers. A dominant negative form of Xenopus T cell-specific factor 3 (XTCF-3) inhibited promoter activity, indicating that T cell-specific factor (TCF)/lymphocyte enhancer binding factor 1 (LEF-1) signaling is necessary for regulation of Siamois. Mutagenesis of two individual TCF sites in the −245 promoter revealed that the proximal, but not distal, site is necessary for dorsovegetal activation. These observations suggest that Siamois is directly regulated by TCFs during dorsoventral axis determination. Further deletion analysis identified a positive regulatory region that is required for dorsal activation, but not for Wnt inducibility, of the promoter. We also present evidence for autoregulation of Siamois transcription. Furthermore, the Siamois promoter was activated by Wnt signaling in 293T tissue culture cells, demonstrating that regulation of the promoter is functionally conserved.
In Xenopus, the dorsoventral axis is specified soon after fertilization, when the symmetry of the egg is broken as a result of cortical rotation. This process involves rearrangement of the egg cytoplasm relative to the cortex and may be mediated by a polarized array of microtubules (1). Cortical rotation is believed to activate cytoplasmic components, called dorsal determinants, along one side of the embryo. This initial asymmetry results in the formation of the organizer in the region of the dorsal blastopore lip at the beginning of gastrulation (1). The organizer is a special cell population that is capable of inducing a secondary body axis upon transplantation to the ventral side of a recipient embryo. As an early signaling center, the organizer directs critical inductive and morphogenetic events required for proper axis formation. A current model for organizer formation involves synergistic interactions between mesoderm-inducing signals, such as transforming growth factor-β and fibroblast growth factors, and dorsal determinants (2).
The nature of the dorsal determinants remains elusive. Overexpression of components of the Wnt pathway, such as certain Wnt ligands, Dishevelled, and β-catenin, can induce an ectopic organizer (3), suggesting that dorsal determinants function through the Wnt pathway. Furthermore, dorsal development is inhibited in embryos lacking maternal β-catenin, but ventral and lateral mesodermal tissues are present (4). β-Catenin may function by entering the nucleus and activating gene transcription in a complex with high mobility group (HMG) box-containing T cell-specific factors (TCF, ref. 5). Overexpression of dominant negative forms of TCF leads to ventralization of Xenopus embryos (6, 7) similar to the effects of β-catenin depletion (4). Together these observations suggest that a β-catenin/TCF complex regulates transcription of critical organizer genes.
The homeobox gene Siamois is one of the earliest genes induced in dorsovegetal cells after the onset of zygotic transcription (8), and its activity is necessary for organizer formation (9, 10). Siamois induces an ectopic axis when overexpressed on the ventral side, and it activates organizer markers in ectodermal explants (8, 11). Siamois can be induced in ectodermal explants by the Wnt signaling pathway in the absence of other inducing factors (11–13). Additionally, functional analysis indicates that Siamois may act downstream of β-catenin (9, 14, 15). Therefore Siamois may be a direct target of β-catenin/TCF signaling in the embryo.
As Siamois appears to be an essential zygotic intermediary between maternal signaling and organizer formation (8–10), examining how Siamois is regulated should yield insight into how the organizer is specified. To this end, we isolated and characterized the upstream regulatory sequences for Siamois. Our analysis demonstrates that correct expression of Siamois requires two promoter regions, one of which contains a TCF consensus site. These regions cooperate to drive transcription of a luciferase reporter gene in a specific spatial and temporal manner, mimicking the endogenous expression of Siamois.
MATERIALS AND METHODS
DNA Constructs.
A 5′-terminal fragment of Siamois cDNA (8) was used to probe a Xenopus genomic λEMBL4 library (a gift of T. Sargent, National Institutes of Health, Bethesda, MD) using standard techniques (16). A 1.05-kb DNA fragment from the phage insert was subcloned into pBluescript SK and sequenced. To prevent an aberrant transcriptional start in promoter-reporter gene constructs, the ATG coding start site of Siamois was mutated to TTG by PCR, using T7 (Promega) and M>L primers (mutated nucleotide underlined, appended BamHI and SacI sites are in boldface): 5′TAGGATCCGAGCTCTAGGTCAAGTCTGTCTCC-3′. The 833-bp fragment of the upstream regulatory sequence of the Siamois promoter was then cloned into pXP2 (17) to generate a fusion with the luciferase coding region (−833pSia-Luc).
Promoter deletions were made using the available restriction sites, S1 exonuclease digestion (18), or PCR with the M>L primer and 5′-GGTAAGCTTCATCAAAGGACCTCCCT-3′. Site-directed mutagenesis of TCF sites was carried out by mutation of an end primer or by PCR overlap extension (19). The following primers were used (mutated nucleotides underlined): (i) 5′-GGTAAGCTTCATCAAGCGACCTCCCT-3′ and M>L primer for −245GC pSia-Luc, and (ii) 5′-CAACAGTACATGGAAGTATTTATA-3′ (upstream) and 5′-TATAAATACTTCCATGTACTGTTG-3′ (downstream) for −296GG pSia-Luc. PCR products were subcloned into pXP2 (17). Mutagenesis was verified by sequencing.
Xenopus Embryos, Microinjections, and Luciferase Assays.
Eggs were obtained from Xenopus females, cultured, and microinjected as described (9). Embryonic stages were determined according to Nieuwkoop and Faber (20). Capped synthetic mRNAs were generated by in vitro transcription of plasmids encoding Xwnt8 (21), a dominant negative form of Xenopus TCF-3 (ΔN-XTCF-3, ref. 6), Xnr3 (22), tBr (23), Siamois (8), and the Siamois-engrailed repressor (SE; ref. 9) using mMessage mMachine kits (Ambion). Each injected blastomere received 10 nl of a solution containing 20 pg of supercoiled pSia-Luc DNA ± 5 pg of Xwnt8, 200 pg of ΔN-XTCF-3, 100 pg of Xnr3, 1 ng of tBr, 3 pg of Siamois, or 10 pg of SE mRNA. Luciferase activity was determined in lysates from stage 10.25–10.5 embryos as described (9). Five embryos were combined for each measurement, and all assays were done in triplicate. Each experiment utilized embryos from the same fertilization batch and was repeated independently at least three times.
Cell Line Transfections.
293T cells were plated in 12-well plates at a density of ≈5 × 104 cells per well 18 hr before transient transfection. Transfection with −833pSia-Luc, a β-galactosidase expression vector, and a vector directing the expression of Xwnt8 (21), Xenopus dishevelled (Xdsh) (24), or β-catenin (25) was performed by the calcium phosphate method (16). Total DNA concentration was 3.66 μg per well. Luciferase activity was determined using the Promega luciferase assay system according to the manufacturer’s protocol, and normalized for β-galactosidase activity.
RESULTS
Upstream Regulatory Sequences Reproduce the Endogenous Expression of Siamois.
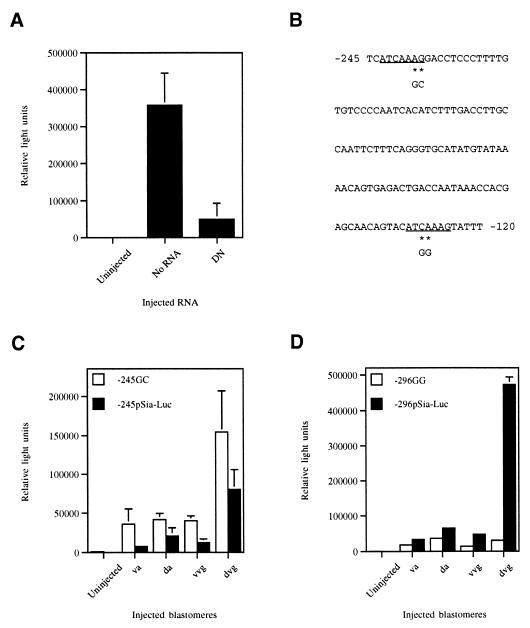
A phage hybridizing with a 5′-terminal fragment of Siamois cDNA was cloned from a Xenopus genomic library. Nucleotide sequencing confirmed that an isolated 1.05-kb insert contained 216 bp of the Siamois coding sequence (8) and 833 bp of 5′-flanking sequences (Fig. 1). There is a TATA-like sequence at position −62 (numbers represent the distance in bp upstream of start codon). Three exact matches to the TCF consensus site CTTTGA/TA/T (26) are present in the promoter sequence at positions −384, −237, and −125. A sequence TTGGTCA similar to the AP1 consensus site was found at position −152 of the promoter. In addition, six basic helix–loop–helix recognition sites, CANNTG, are present at positions −804, −583, −401, −297, −260, and −180.
Figure 1.
Upstream regulatory sequence of the Siamois gene. The beginning of the Siamois coding region and of the corresponding cDNA (italic type) are shown. A TATA-like sequence is underlined. Three TCF consensus binding sites are shown in boldface.
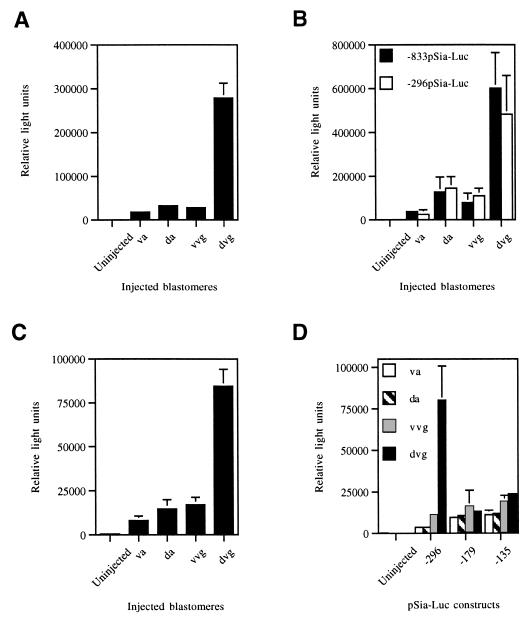
To determine whether the isolated 0.83 kb of 5′-flanking region of the Siamois promoter can activate transcription from a heterologous gene, it was subcloned into the promoterless luciferase reporter plasmid pXP2 (17). The resulting −833pSia-Luc construct was injected into different blastomeres of 4- to 8-cell embryos. Luciferase activity was measured at early gastrula stages (stages 10.25–10.5), when endogenous Siamois transcripts are most abundant (8). Maximal activity was detected when the construct was injected into dorsovegetal blastomeres (Fig. 2A). A control luciferase plasmid driven by a cytomegalovirus promoter was activated equally in different regions of the embryo (data not shown). Because Siamois is known to be expressed in dorsovegetal blastomeres (8), the isolated 5′-flanking sequences closely mimicked regulation of Siamois in vivo.
Figure 2.
Comparison of pSia-Luc constructs in different regions of the embryo. Luciferase activity was measured in embryos microinjected into ventral animal (va), dorsal animal (da), ventral vegetal (vvg), and dorsovegetal (dvg) blastomeres with −833pSia-Luc (A and B), −296pSia-Luc (B and D), −245pSia-Luc (C), and −179pSia-Luc and −135 pSia-Luc (D). Experimental data are expressed as the means from triplicate samples ± SD. One representative experiment is shown in each graph.
Dorsal Activation of Siamois Requires Sequences Between −245 and −179.
To further map the regions of the Siamois promoter that are essential for its activation in the organizer, we generated a series of 5′ deletion constructs fused to the luciferase reporter gene. These constructs were microinjected into early blastomeres and luciferase activity was measured in embryo lysates. The −707pSia-Luc (data not shown), −296pSia-Luc, and −245pSia-Luc constructs were indistinguishable from −833pSia-Luc (Fig. 2 B and C), demonstrating that the proximal 245 bp of the promoter are sufficient to mediate its dorsal activation. However, a considerable loss of activity in dorsovegetal cells was observed for −179pSia-Luc and −135pSia-Luc (Fig. 2D), indicating that the region between −245 and −179 contains essential regulatory element(s) necessary for endogenous expression of Siamois.
The Role for TCF Factors in Regulating Siamois Expression.
To evaluate the role of TCF family members in regulation of the Siamois promoter, the effect of a dominant negative XTCF-3 (ΔN-XTCF-3, ref. 6) on activation of −833pSia-Luc was determined. ΔN-XTCF-3 inhibited the endogenous activation of −833pSia-Luc in dorsovegetal cells (Fig. 3A), indicating that Siamois expression is dependent on TCF-mediated transcriptional activation.
Figure 3.
TCF signaling is critical for Siamois transcription. (A) Activation of −833pSia-Luc in dorsovegetal blastomeres was suppressed by coinjection of ΔN-XTCF-3 (DN) mRNA. (B) Location of mutated TCF consensus sites (underlined) in the promoter. (C and D) Comparison of mutated constructs in various blastomeres. Abbreviations are as in Fig. 2.
To more fully assess the contribution of individual TCF sites in this regulation, we deleted or mutated the TCF consensus sites. Deletion constructs lacking the most distal TCF site at position −384 behaved identically to constructs containing the site (Fig. 2B), demonstrating that the −384 consensus is not essential for dorsovegetal expression of Siamois. Mutation of two bases in the −237 TCF site of −245pSia-Luc (−245GC pSia-Luc) resulted in a ≈3-fold higher level of activity in all blastomeres compared with wild-type −245pSia-Luc (Fig. 3C). In contrast, changing two bases in the proximal −124 TCF site in the −296pSia-Luc construct (−296GG pSia-Luc) dramatically reduced activity in dorsovegetal cells (Fig. 3D). These results reveal that Siamois activation requires the proximal −124 TCF site.
Specific Activation by Wnt Signaling.
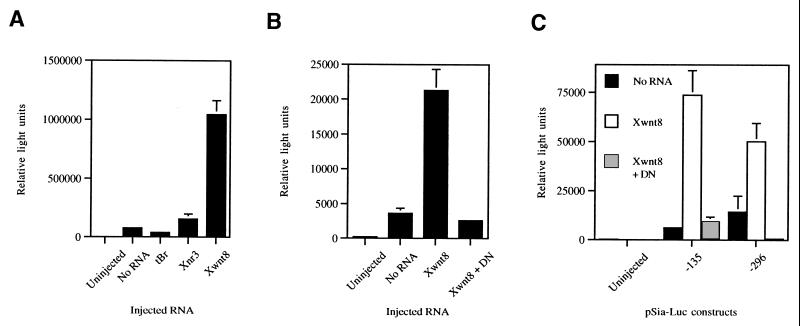
Because Siamois is induced by the Wnt pathway in animal pole cells in the absence of other inducing factors (11, 13), we assessed whether the isolated promoter responded to Wnt signaling. Both animal ventral blastomeres of 4- to 8-cell embryos were injected with −833pSia-Luc or related constructs together with Xwnt8 mRNA. Animal ventral blastomeres have minimal background activity of the Siamois promoter in our assays. Xwnt8, Xwnt2, and β-catenin mRNAs activated −833pSia-Luc ≈5–15-fold (Fig. 4A and data not shown). In contrast, overexpression of Xnr3, a transforming growth factor-β family member (22), did not significantly activate the promoter (Fig. 4A). Interference with transforming growth factor-β signaling by a dominant negative BMP4-receptor (tBR, ref. 23) also failed to affect the promoter (Fig. 4A). Together these findings suggest that Siamois expression is specifically controlled by Wnt signaling.
Figure 4.
Wnt inducibility of the Siamois promoter. Embryos were microinjected in two ventral animal blastomeres with −833pSia-Luc (A), −245pSia-Luc (B), and −135pSia-Luc and −296pSia-Luc (C). Luciferase activity was measured for embryos injected with promoter constructs alone, or coinjected with Xwnt8 ± DN, tBr, or Xnr3 mRNAs as shown.
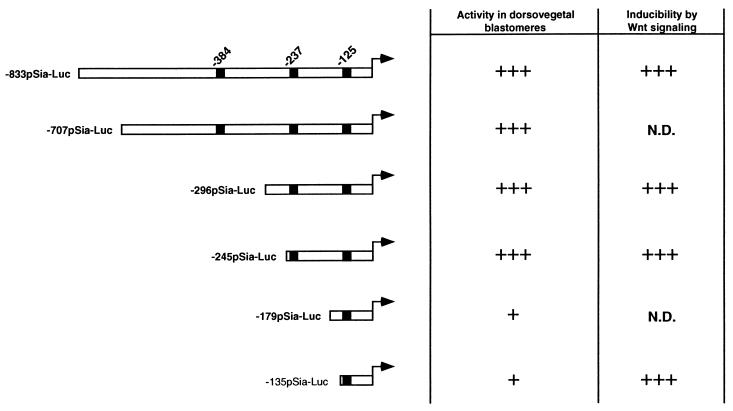
We next tested the Wnt inducibility of other deletion constructs (Fig. 4 B and C; Fig. 5). All tested pSia-Luc plasmids that were active in dorsovegetal blastomeres were similarly responsive to Wnt signaling, and this response was inhibited by coinjection of ΔN-XTCF-3 mRNA (Fig. 4 B and C). While both −245pSia-Luc and −135pSia-Luc were inducible by Wnt signaling, only −245pSia-Luc was activated in dorsovegetal cells (Fig. 2 C and D and Fig. 4 B and C). This indicates that elements located between −245 and −135 are required for induction of the promoter by endogenous signals, but not for activation by the Wnt pathway.
Figure 5.
Activity of different pSia-Luc constructs in dorsovegetal cells and their inducibility by Wnt signaling. Positions of the three TCF consensus sites are shown. N.D. = not determined.
Evidence for Autoregulation of Siamois.
The existence of a positive feedback loop is suggested by the fact that −833pSia-Luc was also stimulated by Siamois mRNA (Fig. 6A). To examine this possibility, a dominant inhibitor of Siamois, SE (9), was coexpressed with various promoter constructs in dorsovegetal cells at the 4- to 8-cell stage. When luciferase activity was analyzed at early gastrula stages, −833pSia-Luc (data not shown), −296pSia-Luc, −245pSia-Luc, and −179pSia-Luc (Fig. 6B) were all inhibited by coinjection of SE mRNA. In contrast, −135pSia-Luc was not inhibited by SE (Fig. 6C). Thus sequences responsible for regulation by SE are located between positions −179 and −135, and they may be involved in maintenance of Siamois expression on the dorsal side.
Figure 6.
Autoregulation of Siamois transcription. (A) Induction of −833pSia-Luc by Siamois mRNA in ventral animal blastomeres. (B and C) The effect of SE mRNA on activity of −296, −245, and −179pSia-Luc constructs (B) and −135pSia-Luc (C) in dorsovegetal blastomeres.
Activation of the Promoter in Mammalian Cells.
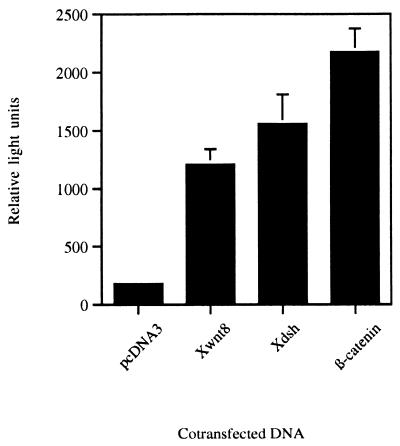
To examine whether the Siamois promoter can respond to Wnt signaling in other systems, we studied its activation in the human embryonic kidney cell line 293T. Use of the Siamois promoter in cell lines may be a useful readout system for studying Wnt signal transduction. Cotransfection with plasmids encoding Xwnt8, Xdsh, or β-catenin resulted in 5–10-fold induction of the luciferase reporter gene as compared with controls (Fig. 7). Thus regulation of the promoter by Wnt signaling is functionally conserved in mammalian cells.
Figure 7.
−833pSia-Luc is activated by Wnt signaling in mammalian cells. 293T cells were transfected with a control pcDNA3 plasmid, or with Xwnt8-pCS2, Xdsh-pCS2, or β-catenin-pcDNA3 plasmids. Luciferase activity is expressed as the means from triplicate samples ± SD.
DISCUSSION
We have shown that 5′-flanking sequences of the Siamois promoter can activate a luciferase reporter gene and mimic endogenous expression of Siamois. Different components of the Wnt pathway were able to specifically activate promoter constructs in Xenopus embryos and in 293T cells, demonstrating functional conservation of the upstream regulatory sequences of Siamois. Mutagenesis studies revealed that transcriptional activation requires two promoter regions, one of which corresponds to a TCF consensus site. The other region is needed for dorsovegetal activation but not for Wnt inducibility. Our data also indicate that Siamois is involved in control of its own transcription. As there are no homeodomain-binding consensus sites (TAAT) within the first 331 bp of 5′ flanking sequence, the observed effects of SE on Siamois transcription are likely to be indirect and may be mediated by other regulatory gene products.
In our study, the most proximal 245 bp are sufficient for complete dorsovegetal activation of the promoter. This is in contrast to the low activity seen with a similar length construct in an independent study (27). The promoter that we cloned contains an additional 14 nucleotides that are not present in the previously reported sequence (27). The sequence disparity may be due to the pseudotetraploid DNA content of Xenopus, and may account for the observed functional differences.
Analysis of the upstream regulatory region of Siamois supports the idea that the function of TCF proteins in Wnt signal transduction is conserved in many species (5). Of the three TCF consensus sites in the cloned promoter, only the most proximal site at −124 appears essential for dorsovegetal activation (Fig. 3D), emphasizing an activating role for TCFs in Siamois regulation. Interestingly, the exact decanucleotide sequence and position of this site are conserved between the promoters of Siamois and the closely related gene Twin (28). These observations suggest a positive role for Wnt signaling in dorsovegetal induction of Siamois.
A negative role for TCF-like transcription factors has been proposed in C. elegans, where the HMG box-containing protein POP-1 appears to work in opposition to Wnt signaling (29, 30). A mutation in the −237 TCF site of the Siamois promoter resulted in 2–3-fold upregulation of activity (Fig. 3C), indicating that this site may mediate repression by TCFs. Similar derepression of the Siamois promoter has been observed in an independent work (27). Together, these and other studies (31) suggest that in some cases, TCFs may function as negative regulators of transcription.
Further analysis of the Siamois promoter revealed essential positive regulatory sequences between −245 and −179 (Fig. 5). This region does not appear to depend on TCF regulation because mutation of the −237 TCF site or of a less conserved site at −208 does not reduce dorsovegetal expression of the promoter constructs (Fig. 3C and data not shown). This region was not required for Wnt inducibility of the Siamois promoter (Fig. 4C). These findings suggest that Siamois transcription depends on factors other than TCFs, and these factors may act in addition to Wnt signaling.
Our data demonstrate that two essential regions of the promoter cooperate to control the endogenous expression of Siamois. Similarly, the Ultrabithorax promoter in Drosophila and the goosecoid promoter in Xenopus have been shown to be regulated by synergistic interactions between Wnt signaling and other pathways (32, 33). Furthermore, although β-catenin is localized to nuclei of both dorsovegetal and dorsoanimal cells in Xenopus blastulae (34, 35), the Siamois promoter is significantly activated only in dorsovegetal blastomeres. Together these studies support a model in which signals other than β-catenin/TCF contribute to the spatial restriction of Siamois expression.
The organizer was proposed to form either by inductive interactions between dorsovegetal and dorsal marginal cells (36), or through direct activation by cytoplasmic determinants (37). The observation that Siamois can be activated in dissociated cells (8, 13) suggests that the organizer may form in the absence of cell-cell interactions. Furthermore, the organizer genes Siamois, Twin, and Xnr3 appear to be directly regulated by β-catenin/TCF complexes (this study; refs. 27, 28, 38). In addition, Twin binds to regulatory regions of goosecoid and activates its transcription (28). Together these findings support a model for cell-autonomous specification of the organizer.
Acknowledgments
We thank T. Sargent for the Xenopus genomic library; O. Destrée, R. Harland, P. Lemaire, P. McCrea, D. Melton, and N. Moghal for plasmids; L. Konnikova for help with experiments; and K. Itoh for critical reading of the manuscript. This work was supported by funding from the Howard Hughes Medical Institute to M.J.F. and by grants from the National Institues of Health and March of Dimes Birth Defects Foundation to S.Y.S.
ABBREVIATIONS
- TCF
T cell-specific factors
- SE
Siamois-engrailed
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF026478).
References
- 1.Gerhart, J., Danilchik, M., Doniach, T., Roberts, S., Rowning, B. & Stewart, R. (1989) Development (Cambridge, U.K.) Suppl., 37–51. [DOI] [PubMed]
- 2.Lemaire P, Kodjabachian L. Trends Genet. 1996;12:525–531. doi: 10.1016/s0168-9525(97)81401-1. [DOI] [PubMed] [Google Scholar]
- 3.Moon R T, Brown J D, Torres M. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 4.Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro C Y, Wylie C. Cell. 1994;79:791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 5.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 6.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destrée O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 7.Behrens J, von Kries J P, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 8.Lemaire P, Garrett N, Gurdon J B. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- 9.Fan M J, Sokol S Y. Development (Cambridge, UK) 1997;124:2581–2589. doi: 10.1242/dev.124.13.2581. [DOI] [PubMed] [Google Scholar]
- 10.Kessler D S. Proc Natl Acad Sci USA. 1997;94:13017–13022. doi: 10.1073/pnas.94.24.13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carnac G, Kodjabachian L, Gurdon J B, Lemaire P. Development (Cambridge, UK) 1996;122:3055–3065. doi: 10.1242/dev.122.10.3055. [DOI] [PubMed] [Google Scholar]
- 12.Yang-Snyder J, Miller J R, Brown J D, Lai C-J, Moon R T. Curr Biol. 1996;6:1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- 13.Brannon M, Kimelman D. Dev Biol. 1996;180:344–347. doi: 10.1006/dbio.1996.0306. [DOI] [PubMed] [Google Scholar]
- 14.Wylie C, Kofron M, Payne C, Anderson R, Hosobuchi M, Joseph E, Heasman J. Development (Cambridge, UK) 1996;122:2987–2996. doi: 10.1242/dev.122.10.2987. [DOI] [PubMed] [Google Scholar]
- 15.Fagotto F, Guger K, Gumbiner B M. Development (Cambridge, UK) 1997;124:453–460. doi: 10.1242/dev.124.2.453. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Nordeen S K. BioTechniques. 1988;6:454–458. [PubMed] [Google Scholar]
- 18.Tabor S, Struhl K. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 1. New York: Greene and Wiley-Interscience; 1989. pp. 3.12.2–3.12.3. [Google Scholar]
- 19.Horton R, Cai Z, Ho S, Pease L. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 20.Nieuwkoop P D, Faber J. Normal Table of Xenopus laevis (Daudin) Amsterdam: North-Holland; 1967. [Google Scholar]
- 21.Christian J L, McMahon J A, McMahon A P, Moon R T. Development (Cambridge, UK) 1991;111:1045–1055. doi: 10.1242/dev.111.4.1045. [DOI] [PubMed] [Google Scholar]
- 22.Smith W C, McKendry R, Ribisi S, Jr, Harland R M. Cell. 1995;82:37–46. doi: 10.1016/0092-8674(95)90050-0. [DOI] [PubMed] [Google Scholar]
- 23.Graff J M, Thies R S, Song J J, Celeste A J, Melton D A. Cell. 1994;79:169–179. doi: 10.1016/0092-8674(94)90409-x. [DOI] [PubMed] [Google Scholar]
- 24.Sokol S Y, Klingensmith J, Perrimon N, Itoh K. Development (Cambridge, UK) 1995;121:1637–1647. doi: 10.1242/dev.121.6.1637. [DOI] [PubMed] [Google Scholar]
- 25.McCrea P D, Turck C W, Gumbiner B M. Science. 1991;254:1359–1361. doi: 10.1126/science.1962194. [DOI] [PubMed] [Google Scholar]
- 26.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 27.Brannon M, Gomperts M, Sumoy L, Moon R T, Kimelman D. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurent M N, Blitz I L, Hashimoto C, Rothbächer U, Cho K W-Y. Development (Cambridge, UK) 1997;124:4905–4916. doi: 10.1242/dev.124.23.4905. [DOI] [PubMed] [Google Scholar]
- 29.Thorpe C J, Schlesinger A, Carter J C, Bowerman B. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- 30.Rocheleau C E, Downs W D, Lin R, Wittmann C, Bei Y, Cha Y-H, Ali M, Priess J R, Mello C C. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- 31.Merriam J M, Rubenstein A B, Klymkowsky M W. Dev Biol. 1997;185:67–81. doi: 10.1006/dbio.1997.8550. [DOI] [PubMed] [Google Scholar]
- 32.Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S-C, Grosschedl R, Bienz M. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 33.Watabe T, Kim S, Candia A, Rothbächer U, Hashimoto C, Inoue K, Cho K W-Y. Genes Dev. 1995;9:3038–3050. doi: 10.1101/gad.9.24.3038. [DOI] [PubMed] [Google Scholar]
- 34.Schneider S, Steinbeisser H, Warga R M, Hausen P. Mech Dev. 1996;57:191–198. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- 35.Larabell C A, Torres M, Rowning B A, Yost C, Miller J R, Wu M, Kimelman D, Moon R T. J Cell Biol. 1997;136:1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gimlich R L, Gerhart J C. Dev Biol. 1984;104:117–130. doi: 10.1016/0012-1606(84)90042-3. [DOI] [PubMed] [Google Scholar]
- 37.Yuge M, Kobayakawa Y, Fujisue M, Yamana K. Development (Cambridge, UK) 1990;110:1951–1956. doi: 10.1242/dev.110.4.1051. [DOI] [PubMed] [Google Scholar]
- 38.McKendry R, Hsu S-C, Harland R M, Grosschedl R. Dev Biol. 1997;192:420–431. doi: 10.1006/dbio.1997.8797. [DOI] [PubMed] [Google Scholar]