Abstract
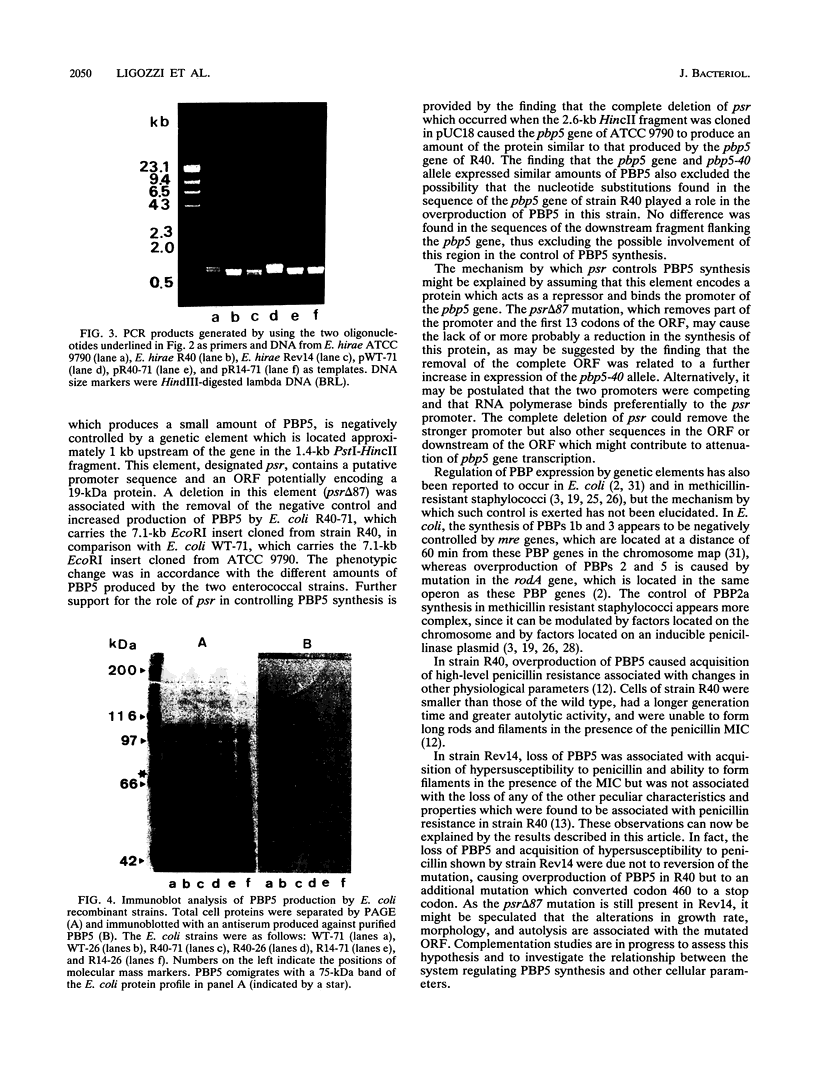
Enterococcus hirae ATCC 9790 produces a penicillin-binding protein (PBP5) of low penicillin affinity which under certain conditions can take over the functions of all the other PBPs. The 7.1-kb EcoRI fragment containing the pbp5 gene of this strain and of two mutants, of which one (E. hirae R40) overproduces PBP5 and the other (E. hirae Rev14) does not produce PBP5, was cloned in pUC18 and sequenced. In the 7.1-kb EcoRI fragment cloned from strain ATCC 9790, an open reading frame (psr) potentially encoding a 19-kDa protein was identified 1 kb upstream of the pbp5 gene. An 87-bp deletion in this element was found in the 7.1-kb EcoRI fragment cloned from strains R40 and Rev14. In addition, several base substitutions were found in the pbp5 genes of strains R40 and Rev14. One of these converted the 42nd codon, TCA, to the stop codon, TAA, in the pbp5 gene of Rev14. Escherichia coli strains were transformed with plasmids carrying the 7.1-kb EcoRI insert or a 2.6-kb HincII insert containing only the pbp5 gene of the three strains. Immunoblotting analysis of proteins expressed by these transformants showed that the 87-bp deletion in psr was associated with the PBP5 overproducer phenotype of strain R40 and the conversion of the TCA codon to the stop codon was associated with the PBP5 nonproducer phenotype of strain Rev14. None of the other nucleotide substitutions had any apparent effect on the level of PBP5 synthesized.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Begg K. J., Takasuga A., Edwards D. H., Dewar S. J., Spratt B. G., Adachi H., Ohta T., Matsuzawa H., Donachie W. D. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J Bacteriol. 1990 Dec;172(12):6697–6703. doi: 10.1128/jb.172.12.6697-6703.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Strässle A., Gustafson J. E., Kayser F. H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1992 Jul;36(7):1367–1373. doi: 10.1128/aac.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. F., Reynolds P. E. Intrinsic resistance to beta-lactam antibiotics in Staphylococcus aureus. FEBS Lett. 1980 Dec 29;122(2):275–278. doi: 10.1016/0014-5793(80)80455-8. [DOI] [PubMed] [Google Scholar]
- Canepari P., Lleò M. M., Cornaglia G., Fontana R., Satta G. In Streptococcus faecium penicillin-binding protein 5 alone is sufficient for growth at sub-maximal but not at maximal rate. J Gen Microbiol. 1986 Mar;132(3):625–631. doi: 10.1099/00221287-132-3-625. [DOI] [PubMed] [Google Scholar]
- Canepari P., Lleò M. M., Fontana R., Satta G. Streptococcus faecium mutants that are temperature sensitive for cell growth and show alterations in penicillin-binding proteins. J Bacteriol. 1987 Jun;169(6):2432–2439. doi: 10.1128/jb.169.6.2432-2439.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers H. F., Hartman B. J., Tomasz A. Increased amounts of a novel penicillin-binding protein in a strain of methicillin-resistant Staphylococcus aureus exposed to nafcillin. J Clin Invest. 1985 Jul;76(1):325–331. doi: 10.1172/JCI111965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyette J., Ghuysen J. M., Fontana R. The penicillin-binding proteins in Streptococcus faecalis ATCC 9790. Eur J Biochem. 1980 Sep;110(2):445–456. doi: 10.1111/j.1432-1033.1980.tb04886.x. [DOI] [PubMed] [Google Scholar]
- Fontana R., Canepari P., Satta G., Coyette J. Identification of the lethal target of benzylpenicillin in Streptococcus faecalis by in vivo penicillin binding studies. Nature. 1980 Sep 4;287(5777):70–72. doi: 10.1038/287070a0. [DOI] [PubMed] [Google Scholar]
- Fontana R., Canepari P., Satta G., Coyette J. Streptococcus faecium ATCC 9790 penicillin-binding proteins and penicillin sensitivity are heavily influenced by growth conditions: proposal for an indirect mechanism of growth inhibition by beta-lactams. J Bacteriol. 1983 May;154(2):916–923. doi: 10.1128/jb.154.2.916-923.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana R., Cerini R., Longoni P., Grossato A., Canepari P. Identification of a streptococcal penicillin-binding protein that reacts very slowly with penicillin. J Bacteriol. 1983 Sep;155(3):1343–1350. doi: 10.1128/jb.155.3.1343-1350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana R., Grossato A., Rossi L., Cheng Y. R., Satta G. Transition from resistance to hypersusceptibility to beta-lactam antibiotics associated with loss of a low-affinity penicillin-binding protein in a Streptococcus faecium mutant highly resistant to penicillin. Antimicrob Agents Chemother. 1985 Nov;28(5):678–683. doi: 10.1128/aac.28.5.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Nagata K., Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase: effect of base substitutions in the promoter -35 region on promoter strength. Nucleic Acids Res. 1990 Dec 25;18(24):7367–7372. doi: 10.1093/nar/18.24.7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ligozzi M., Aldegheri M., Predari S. C., Fontana R. Detection of penicillin-binding proteins immunologically related to penicillin-binding protein 5 of Enterococcus hirae ATCC 9790 in Enterococcus faecium and Enterococcus faecalis. FEMS Microbiol Lett. 1991 Oct 15;67(3):335–339. doi: 10.1016/0378-1097(91)90498-y. [DOI] [PubMed] [Google Scholar]
- Murakami K., Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol. 1989 Feb;171(2):874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras G., el Kharroubi A., van Beeumen J., Coeme E., Coyette J., Ghuysen J. M. Characterization of an Enterococcus hirae penicillin-binding protein 3 with low penicillin affinity. J Bacteriol. 1990 Dec;172(12):6856–6862. doi: 10.1128/jb.172.12.6856-6862.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Predari S. C., Ligozzi M., Fontana R. Genotypic identification of methicillin-resistant coagulase-negative staphylococci by polymerase chain reaction. Antimicrob Agents Chemother. 1991 Dec;35(12):2568–2573. doi: 10.1128/aac.35.12.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L., Tonin E., Cheng Y. R., Fontana R. Regulation of penicillin-binding protein activity: description of a methicillin-inducible penicillin-binding protein in Staphylococcus aureus. Antimicrob Agents Chemother. 1985 May;27(5):828–831. doi: 10.1128/aac.27.5.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. D., Wachi M., Doi M., Ishino F., Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987 Aug 31;221(1):167–171. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- Tesch W., Ryffel C., Strässle A., Kayser F. H., Berger-Bächi B. Evidence of a novel staphylococcal mec-encoded element (mecR) controlling expression of penicillin-binding protein 2'. Antimicrob Agents Chemother. 1990 Sep;34(9):1703–1706. doi: 10.1128/aac.34.9.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trees D. L., Iandolo J. J. Identification of a Staphylococcus aureus transposon (Tn4291) that carries the methicillin resistance gene(s). J Bacteriol. 1988 Jan;170(1):149–154. doi: 10.1128/jb.170.1.149-154.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata K., Nonoguchi R., Matsuhashi M., Konno M. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J Bacteriol. 1989 May;171(5):2882–2885. doi: 10.1128/jb.171.5.2882-2885.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata K., Nonoguchi R., Song M. D., Matsuhashi M., Konno M. Homology of mecA gene in methicillin-resistant Staphylococcus haemolyticus and Staphylococcus simulans to that of Staphylococcus aureus. Antimicrob Agents Chemother. 1990 Jan;34(1):170–172. doi: 10.1128/aac.34.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata K., Yamashita N., Konno M. Occurrence of a beta-lactam-inducible penicillin-binding protein in methicillin-resistant staphylococci. Antimicrob Agents Chemother. 1985 May;27(5):851–857. doi: 10.1128/aac.27.5.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M., Matsuhashi M. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J Bacteriol. 1989 Jun;171(6):3123–3127. doi: 10.1128/jb.171.6.3123-3127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson R., le Bouguénec C., Gutmann L., Horaud T. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to benzylpenicillin. J Gen Microbiol. 1985 Aug;131(8):1933–1940. doi: 10.1099/00221287-131-8-1933. [DOI] [PubMed] [Google Scholar]
- al-Obeid S., Gutmann L., Williamson R. Modification of penicillin-binding proteins of penicillin-resistant mutants of different species of enterococci. J Antimicrob Chemother. 1990 Nov;26(5):613–618. doi: 10.1093/jac/26.5.613. [DOI] [PubMed] [Google Scholar]
- el Kharroubi A., Jacques P., Piras G., Van Beeumen J., Coyette J., Ghuysen J. M. The Enterococcus hirae R40 penicillin-binding protein 5 and the methicillin-resistant Staphylococcus aureus penicillin-binding protein 2' are similar. Biochem J. 1991 Dec 1;280(Pt 2):463–469. [PMC free article] [PubMed] [Google Scholar]