Abstract
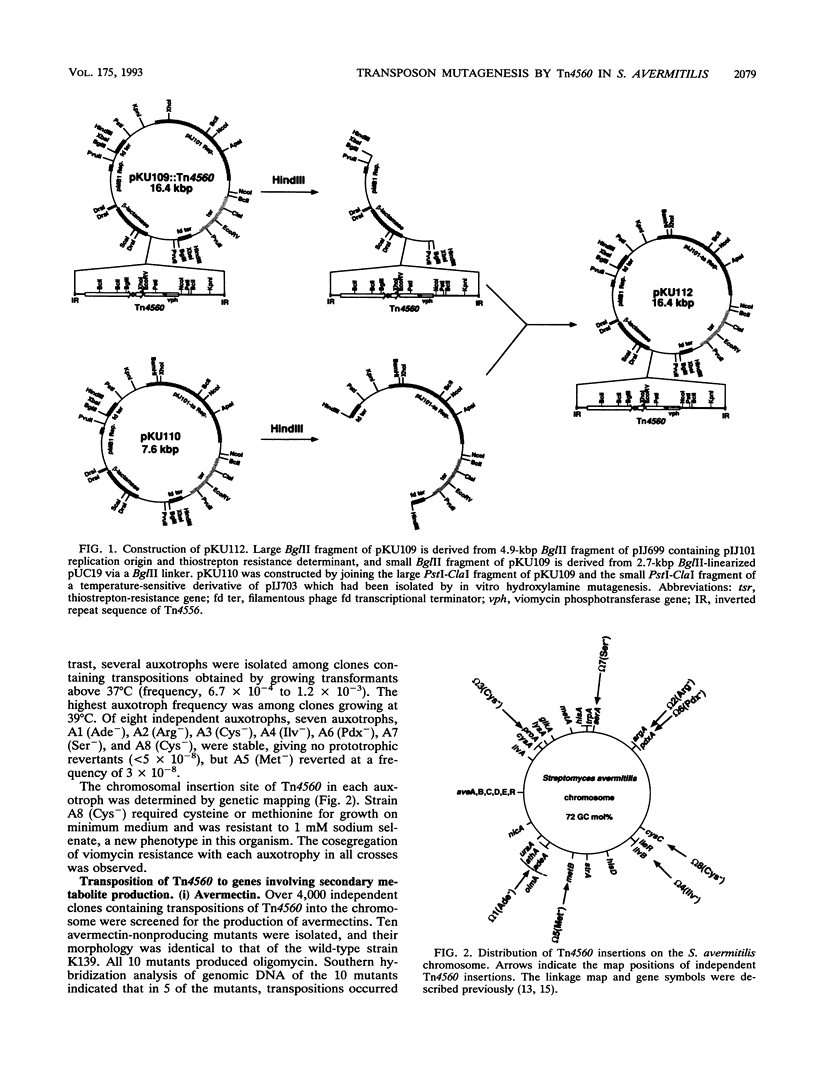
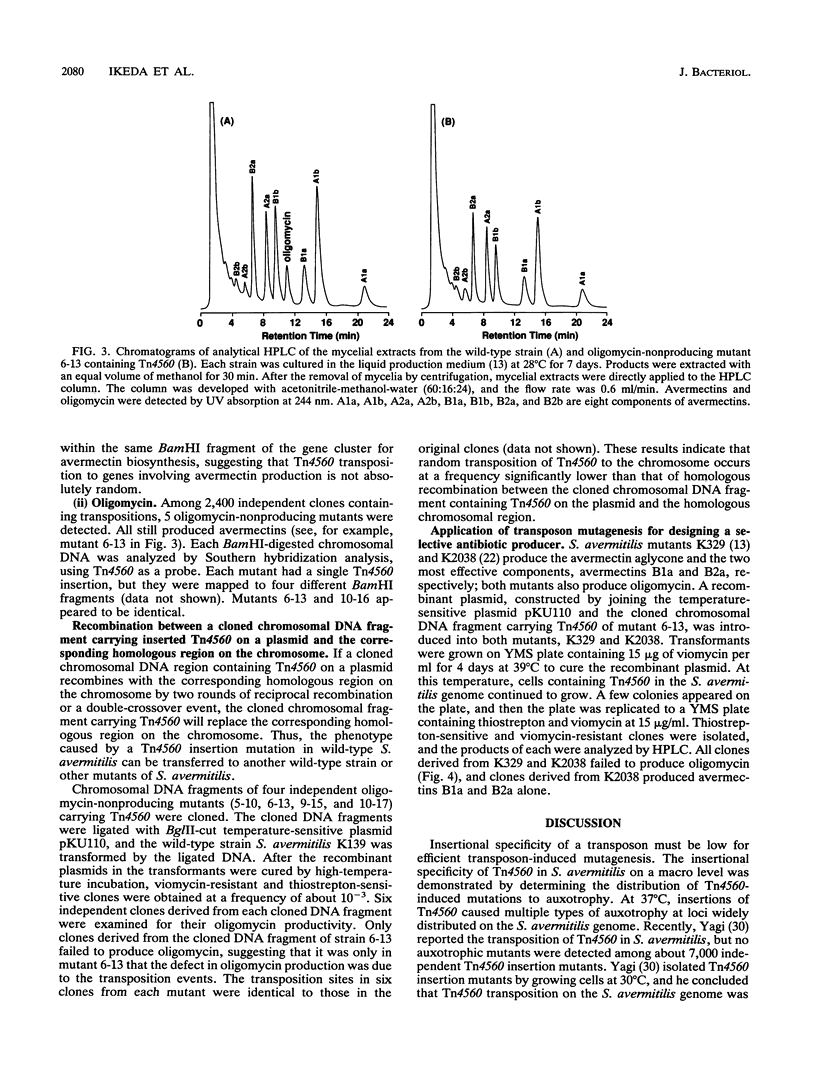
The Tn3-like Streptomyces transposon Tn4560 was used to mutagenize Streptomyces avermitilis, the producer of anthelmintic avermectins and the cell growth inhibitor oligomycin. Tn4560 transposed in this strain from a temperature-sensitive plasmid to the chromosome and from the chromosome to a plasmid with an apparent frequency of about 10(-4) to 10(-3) at both 30 and 39 degrees C. Auxotrophic and antibiotic nonproducing mutations were, however, obtained only with cultures that were kept at 37 or 39 degrees C. About 0.1% of the transposon inserts obtained at 39 degrees C caused auxotrophy or abolished antibiotic production. The sites of insertion into the S. avermitilis chromosome were mapped. Chromosomal DNA fragments containing Tn4560 insertions in antibiotic production genes were cloned onto a Streptomyces plasmid with temperature-sensitive replication and used to transport transposon mutations to other strains, using homologous recombination. This technique was used to construct an avermectin production strain that no longer makes the toxic oligomycin.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burg R. W., Miller B. M., Baker E. E., Birnbaum J., Currie S. A., Hartman R., Kong Y. L., Monaghan R. L., Olson G., Putter I. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother. 1979 Mar;15(3):361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S. T. Tn4556, a 6.8-kilobase-pair transposable element of Streptomyces fradiae. J Bacteriol. 1987 Oct;169(10):4436–4441. doi: 10.1128/jb.169.10.4436-4441.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerick A. W. Read-through transcription from a derepressed Tn3 promoter affects ColE1 functions on a ColE1::Tn3 composite plasmid. Mol Gen Genet. 1982;185(3):408–417. doi: 10.1007/BF00334132. [DOI] [PubMed] [Google Scholar]
- Harayama S., Tsuda M., Iino T. Tn1 insertion mutagenesis in Escherichia coli K-12 using a temperature-sensitive mutant of plasmid RP4. Mol Gen Genet. 1981;184(1):52–55. doi: 10.1007/BF00271194. [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Sekiguchi M. Isolation of temperature-sensitive mutants of R plasmid by in vitro mutagenesis with hydroxylamine. J Bacteriol. 1976 Sep;127(3):1561–1563. doi: 10.1128/jb.127.3.1561-1563.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Kotaki H., Omura S. Genetic studies of avermectin biosynthesis in Streptomyces avermitilis. J Bacteriol. 1987 Dec;169(12):5615–5621. doi: 10.1128/jb.169.12.5615-5621.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Kotaki H., Tanaka H., Omura S. Involvement of glucose catabolism in avermectin production by Streptomyces avermitilis. Antimicrob Agents Chemother. 1988 Feb;32(2):282–284. doi: 10.1128/aac.32.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984 Jul;12(1):19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Kieser T., Melton R. E. Plasmid pIJ699, a multi-copy positive-selection vector for Streptomyces. Gene. 1988 May 15;65(1):83–91. doi: 10.1016/0378-1119(88)90419-2. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Meade H. M., Long S. R., Ruvkun G. B., Brown S. E., Ausubel F. M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982 Jan;149(1):114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura S., Ikeda H., Tanaka H. Selective production of specific components of avermectins in Streptomyces avermitilis. J Antibiot (Tokyo) 1991 May;44(5):560–563. doi: 10.7164/antibiotics.44.560. [DOI] [PubMed] [Google Scholar]
- Pinna L. A., Lorini M., Moret V., Siliprandi N. Effect of oligomycin and succinate on mitochondrial metabolism of adenine nucleotides. Biochim Biophys Acta. 1967 Jul 5;143(1):18–25. doi: 10.1016/0005-2728(67)90105-3. [DOI] [PubMed] [Google Scholar]
- Sandman K., Losick R., Youngman P. Genetic analysis of Bacillus subtilis spo mutations generated by Tn917-mediated insertional mutagenesis. Genetics. 1987 Dec;117(4):603–617. doi: 10.1093/genetics/117.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemieniak D. R., Slightom J. L., Chung S. T. Nucleotide sequence of Streptomyces fradiae transposable element Tn4556: a class-II transposon related to Tn3. Gene. 1990 Jan 31;86(1):1–9. doi: 10.1016/0378-1119(90)90107-3. [DOI] [PubMed] [Google Scholar]
- Solenberg P. J., Baltz R. H. Transposition of Tn5096 and other IS493 derivatives in Streptomyces griseofuscus. J Bacteriol. 1991 Feb;173(3):1096–1104. doi: 10.1128/jb.173.3.1096-1104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solenberg P. J., Burgett S. G. Method for selection of transposable DNA and characterization of a new insertion sequence, IS493, from Streptomyces lividans. J Bacteriol. 1989 Sep;171(9):4807–4813. doi: 10.1128/jb.171.9.4807-4813.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M., Harayama S., Iino T. Tn501 insertion mutagenesis in Pseudomonas aeruginosa PAO. Mol Gen Genet. 1984;196(3):494–500. doi: 10.1007/BF00436198. [DOI] [PubMed] [Google Scholar]
- Weinstock G. M., Susskind M. M., Botstein D. Regional specificity of illegitimate recombination by the translocatable ampicillin-resistance element Tn1 in the genome of phage P22. Genetics. 1979 Jul;92(3):685–710. doi: 10.1093/genetics/92.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T. Transposition of Tn4560 in Streptomyces avermitilis. J Antibiot (Tokyo) 1990 Sep;43(9):1204–1205. doi: 10.7164/antibiotics.43.1204. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Youngman P. J., Perkins J. B., Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]



