Abstract
The gene TP53, encoding transcription factor p53, is mutated or deleted in half of human cancers, demonstrating the crucial role of p53 in tumor suppression. Importantly, p53 inactivation in cancers can also result from the amplification / overexpression of its specific inhibitors MDM2 and MDM4 (also known as MDMX). The presence of wild-type p53 in those tumors with MDM2 or MDM4 overexpression stimulates the search for new therapeutic agents to selectively reactivate it. This short survey highlights recent insights into MDM2 and MDM4 regulatory functions and their implications for the design of future p53-based anticancer strategies. We now know that MDM2 and MDM4 inhibit p53 in distinct and complementary ways: MDM4 regulates p53 activity, while MDM2 mainly regulates p53 stability. Upon DNA damage, MDM2-dependent degradation of itself and MDM4 contribute significantly to p53 stabilization and activation. These and other data imply that the combined use of MDM2 and MDM4 antagonists in cancer cells expressing wild type p53 should activate p53 more significantly than agents that only antagonize MDM2, resulting in more effective anti-tumor activity.
Keywords: MDM2, MDM4, MDMX, p53
1. Introduction
The murine double minute 2 (MDM2) gene was identified in the late 1980s as one of three unknown genes (MDM1-3) coamplified in the spontaneously transformed 3T3-DM mouse cell line. Within five years, the oncogenic potential of MDM2 was demonstrated, the MDM2 protein was shown to bind and inhibit p53, and the human gene homolog (also called MDM2 or HDM2) was found amplified in human sarcomas. Amplification of the MDM2 gene is now reported in more than 10% of 8000 human cancers from various sites, including lung or stomach (reviewed in Toledo and Wahl, 2006). Furthermore, a single nucleotide polymorphism in the MDM2 promoter that leads to a 2-3 fold increase in MDM2 expression correlates with accelerated tumor formation (for review, see Levine et al., 2006).
In the mid 90s, a screen for p53-binding proteins led to the identification of a protein sharing structural homology with MDM2 (reviewed in Marine et al., 2007). The protein was first called MDMX, and later given the official name MDM4 (the human homolog is known as MDM4, MDMX, HDM4 or HDMX). The importance of MDM4 in human cancers has emerged in the past 2 years: MDM4 was found amplified or overexpressed in 10-20% of over 800 diverse tumors including lung, colon, stomach and breast cancers (Toledo and Wahl, 2006) and, strikingly, 65% of retinoblastomas (Laurie et al., 2006).
The regulation of the p53 pathway is proposed to occur through an interaction of p53 with numerous proteins, more than 160 to this date (Toledo and Wahl, 2006) - and a new p53-binding protein is described almost every month. Among these, MDM2 and MDM4 stand out because, in addition to their frequent altered expression in cancers, they were shown to act as essential and specific p53 inhibitors during embryonic development. Indeed, both MDM2-deficient and MDM4-deficient mice die in utero, but these deficiencies are viable in a p53-deficient background (reviewed in Marine et al., 2006). These early mouse studies also indicated that MDM2 and MDM4 are non-redundant p53 inhibitors, as normal levels of either regulator cannot compensate for the loss of the other. Additional in vivo studies that provide insight into the MDM2-MDM4-p53 regulatory network are reviewed below.
2. Protein Structures
Human MDM2 and MDM4 are structurally related proteins of 491 and 490 amino acids respectively, with three well-conserved domains: an N-terminal domain important for binding to the N-terminal part of p53, a Zinc-finger domain (which function remains largely unknown), and a C-terminal RING domain. Both proteins also contain a region rich in acidic residues, without any significant sequence conservation however (Figure 1).
Figure 1.
Comparison of MDM2 and MDM4 primary structures. The p53-BoxI binding domain (BoxI BD; amino acids ca. 25-110), the Zinc finger domain (ZD; aa ca. 290-330) and the RING domain (RING; aa ca. 435-482) are conserved. The BoxI BD is the most conserved domain, and a sequence comparison of amino acids most important for interaction with p53 are shown, with residues that constitute the p53-binding hydrophobic pocket in bold (see text for details). A « lid » before the p53-BoxI BD (i; aa 16-24), which sequence is not conserved, is also proposed to regulate interactions with p53. Both proteins contain a region rich in acidic residues (Acid; aa 237-288 in MDM2, aa 215-255 in MDM4), but these regions do not share any significant sequence homology. The Acidic region in MDM2 is proposed to interact with the S9-S10 β sheets and BoxV from the p53 DNA binding domain, and is thus noted BV BD. L, nuclear localization signal; E, nuclear export signal.
The binding between the N-terminal domain of MDM2 and the N-terminal domain of p53 has been analyzed by X-ray crystallography (Chene, 2004, for review). Residues 15-29 of p53 are part of a highly conserved region (commonly called BoxI). As this region is important for interaction with the basal transcription machinery and transcriptional co-activators, it is also called the p53 transactivation domain (TAD). The p53 residues 15-29 do not appear to adopt a stably folded structure in solution, but residues 19-25 form an α-helix when bound to MDM2. The interaction between p53 and MDM2 is essentially hydrophobic: p53 residues F19 and W23 are located face to face on the same side of the α-helix and, together with p53 L26, they point toward a cleft at the surface of the MDM2 protein, where they are surrounded by hydrophobic MDM2 residues L54, L57, I61, M62, Y67, V75, F86, F91, V93, I99, Y100 and I103. Furthermore, p53-MDM2 interactions are stabilized by intermolecular H-bonds between p53 F19 and MDM2 Q72, p53 W23 and MDM2 L54, and p53 N29 and MDM2 Y100. Thus, 13 residues in the MDM2 p53 BoxI-binding domain appear particularly important for p53 interactions (Figure 1). Importantly, 10 out of these 13 residues are conserved in MDM4, so that the cleft at the surface of MDM4 is similar to, but not identical with, that of MDM2. Furthermore, a flexible « lid » in MDM2 also regulates p53-MDM2 interactions, and the sequence for this lid is very different in MDM4 (Figure 1). Together, these observations suggest subtle but significant differences in the structure and regulation of p53-MDM2 and p53-MDM4 interfaces. Furthermore, recent studies have indicated a more complex regulation for p53-MDM2 interactions: the binding of the N-terminal domain of MDM2 with p53 BoxI may promote conformational changes in MDM2 to stabilize interaction of the MDM2 acidic domain with the p53 DNA binding domain (in a p53 region comprising β-sheets S9-S10 and the conserved BoxV; Wallace et al., 2006). Whether or not the MDM4 acidic region may also interact with the p53 DNA binding domain is presently unknown.
The C-terminal RING domains of MDM2 and MDM4 are essential for these proteins to form homo- or hetero-dimers. Heterodimerization was proposed to be more stable than homodimerization of each protein. Importantly however, such a proposal results from yeast-two hybrid assays and may not directly apply to physiological conditions in mammalian cells. Indeed, the ability of MDM2 and MDM4 to homo- or hetero-dimerize may depend on their relative intracellular concentrations, which are likely to vary depending on growth conditions (see below, Biological functions). An important difference between MDM2 and MDM4 is that the RING domain of MDM2 is essential for its action as an E3-ubiquitin ligase, whereas MDM4 apparently has no intrinsic ubiquitin-ligase activity (see below, Biological Functions).
Finally, as is the case for p53, several amino acid residues in MDM2 and MDM4 can be phosphorylated, ubiquitinated, sumoylated or acetylated, and in vitro studies suggest that these modifications affect MDM2 and MDM4 function and stability. Importantly however, mouse mutants with targeted mutations of specific modified p53 residues often exhibit more modest phenotypes than suggested from in vitro studies (Toledo and Wahl, 2006), indicating that mutations of modified MDM2 and MDM4 residues will have to be analyzed in vivo to precisely evaluate the functional importance of their modifications.
3. Biological Functions
MDM2 has long been considered a major p53 regulator. In vitro and transfection studies led to the conclusion that MDM2 inhibits p53 in two major ways: 1) by interacting with p53 BoxI, thereby preventing p53 interactions with the basal transcription machinery and transcriptional co-activators such as p300; and 2) by regulating p53 stability through ubiquitin-mediated proteasomal degradation. This dual regulation would itself be regulated by a negative feedback loop, as the MDM2 gene is a target for p53-mediated transactivation. The notion that MDM2 is a major p53 regulator was confirmed in vivo when MDM2-deficient embryos were found to die around 4 days post-coitum (dpc) from p53-dependent apoptosis (reviewed in Marine et al., 2006)).
In the p53 regulation model above however, MDM4 did not play an important role. In vitro studies indicated that MDM4 is not a p53 target gene, and that the MDM4 protein may participate in inhibiting p53 activity by interacting with p53 BoxI, but that it lacks intrinsic ubiquitin-ligase activity. The observation that MDM4-deficient embryos die around 10 dpc, apparently from p53-dependent cell proliferation arrest, showed that MDM4 is also an essential p53 regulator (Marine et al., 2006). Additional transfection studies then led to the proposal that a deficiency in either regulator is lethal because of a mutual dependency of these inhibitors, as MDM4 was envisioned to stabilize MDM2, and MDM2 was hypothesized to enable the nuclear import of MDM4 (reviewed in Toledo and Wahl, 2006)).
However, several in vivo approaches provide data that are not consistent with these hypotheses and instead indicate that MDM2 and MDM4 have distinct but complementary roles in p53 regulation. Evidence for this was obtained when a mouse mutant expressing a p53 deleted for its proline-rich domain (p53ΔP) was analyzed (Toledo et al., 2006). We found that p53ΔP is a hypomorphic protein, with deficient cell cycle arrest and reduced apoptotic responses, which allows survival of MDM4-deficient, but not MDM2-deficient animals. The analysis of p53ΔP/ΔP MDM4-/- cells then showed that MDM4 loss does not alter MDM2 stability, but that it increases p53ΔP transactivation capacity significantly, to partially restore a cycle arrest response. Importantly, as MDM2 is a p53 target gene, MDM4 loss also led to higher MDM2 levels and a more efficient degradation of p53ΔP. By contrast, the loss of one MDM2 gene copy led to increased p53ΔP levels, but did not alter, on a per molecule basis, the activity of p53ΔP. This led us to conclude that MDM2 mainly regulates p53 stability and that MDM4 has a major role in regulating p53 activity (Toledo et al., 2006). Another approach, which relied on a Cre-induced conditional activation of p53 in cells deficient in MDM2 or MDM4 led to a similar conclusion (Francoz et al., 2006), and MDM2 and MDM4 were aso shown to have distinct but cooperating roles in the developing CNS (Xiong et al., 2006).
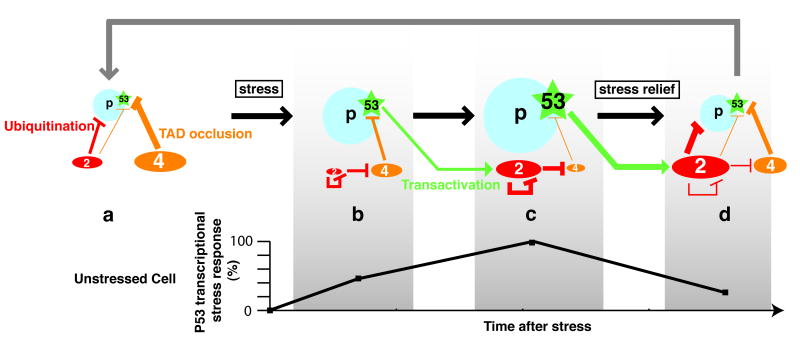
The surprising finding that decreasing MDM2 does not appear to increase p53 activity can be explained by recent studies indicating that DNA damage induces MDM2 self-degradation and an MDM2-dependent degradation of MDM4, a process essential to mount a p53 response. These data suggest a dynamic model for the p53 response that integrates the disctinct and complementary roles of MDM2 and MDM4 in p53 inhibition, and the role of MDM2-mediated degradation of itself and MDM4 for p53 activation. The model is presented in detail in Figure 2. How MDM2 switches from p53 as a degradation target in unstressed cells to ubiquitination of itself and MDM4 after DNA damage remains to be precisely determined, but recent reports provide some clues on the regulation of this switch. The deubiquitinating enzyme Herpes virus-associated ubiquitin-specific protease (HAUSP) has been shown to interact with and stabilize MDM2, MDM4 and p53. Structural studies indicate that both MDM2 and p53 can bind the same site on HAUSP, suggesting that they may compete with each other for this interaction (Sheng et al., 2006). In unstressed cells, the Death-domain associated protein (Daxx) would associate with both MDM2 and HAUSP, and the formation of this complex would reduce MDM2 auto-ubiquitination, enabling MDM2-p53 interactions, and thus p53 degradation. DNA damage would disrupt the MDM2-Daxx-HAUSP complex, resulting in increased MDM2 degradation and p53 stabilization. These findings suggest that Daxx directs HAUSP toward its associated protein MDM2, to increase MDM2 activity towards p53 (Tang et al., 2006a). Daxx-MDM2 interactions may also regulate MDM4 stability (Ronai, 2006), through mechanisms that remain uncertain. HAUSP was also recently proposed to regulate p53-dependent mitochondrial apoptosis (Marchenko et al., 2007). Thus, HAUSP and Daxx emerge as important components of the p53-MDM2-MDM4 regulatory network.
Figure 2.
A dynamic model of the p53 response integrating the distinct and complementary roles of MDM2 and MDM4. (a) in unstressed cells p53 is kept at low levels (due to MDM2-mediated degradation) and inactive (primarily due to MDM4-mediated occlusion of the p53 TAD). In this diagram, p53 stability is represented by a blue circle and p53 activity by a green star, and MDM2 (2) and MDM4 (4) levels are represented by red and orange ovals, respectively. (b) after stress, MDM2 degrades itself and MDM4: a transcriptional stress response (diagrammed below) is mounting. (c) increased MDM2 levels, resulting from p53 activation, lead to a more efficient MDM4 degradation, enabling full p53 activation: the transcriptional response is maximal. (d) following stress relief, accumulated MDM2 targets p53 again, and as MDM4 levels also increase, p53 activity decreases, so that the stress response is fading. This may allow cell cycle re-entry (grey arrow). As discussed in the text, the switch that makes MDM2 preferentially target p53 for degradation in unstressed cells (a), then target itself and MDM4 after stress (b and c) then target p53 again after stress relief (d) is proposed to result from the regulated deubiquitination of p53, MDM2 and MDM4 by HAUSP, a process also involving the adaptor protein Daxx. The model is adapted from (Toledo et al., 2006).
The regulation model in Figure 2 may explain differences in survival of MDM2 and MDM4-deficient embryos in a p53ΔP context (Toledo et al., 2006). Similarly, it may also explain the differences in timing and causes of death in MDM2-null and MDM4-null embryos expressing wild-type p53: in MDM2-deficient embryos, p53 would rapidly accumulate to extremely high levels that exceed MDM4, resulting in continuous activation of a plethora of cell cycle arrest and apoptotic p53 target genes to rapidly induce lethality. In addition, extremely high p53 levels in the cytoplasm may participate in inducing apoptosis via mitochondrial membrane permeabilization (Chipuk and Green, 2006). In MDM4-deficient embryos however, p53 would be more active, but increased MDM2 levels would efficiently degrade it, and moderate its transactivation activity. In this case, low p53 levels in the cytoplasm are unlikely to induce mitochondrial membrane permeabilization, and low p53 levels in the nucleus could titrate p53 binding to chromatin, to restrict its interaction with only those promoters to which it has the highest affinity. Under these conditions, a cell proliferation arrest could be favored in most tissues because p53 apparently binds the promoter of the p21 gene (a major determinant of proliferation arrest) much more efficiently than promoters of many pro-apoptotic genes (Knights et al., 2006). A nonexclusive possibility is that variations in p53 levels may affect its interaction with other proteins proposed to promote cell cycle arrest (e.g. E4F1; LeCam et al., 2006) or apoptosis (e.g. ASPP proteins or TIP60; Sykes et al., 2006; Tang et al., 2006b; Trigiante and Lu, 2006), thereby leading to different responses.
Results consistent with the regulation model in Figure 2 were obtained in independent in vivo studies, in cell types including fibroblasts and neural progenitors (Toledo et al., 2006; Francoz et al., 2006). A recent analysis of the p53 response to ribosomal stress in HCT116 and U2OS cells is also consistent with this model (Gilkes et al., 2006). Thus, this regulation model applies to many tissues, although it remains possible that the relative contribution of MDM2 and MDM4 to p53 regulation varies in some tissues or during specific differentiation phases (discussed in (Marine et al., 2006; Boesten et al., 2006; Maetens et al., 2007). Importantly, a direct implication of the regulation model is that p53 stability and activity are regulated in distinct ways, a notion further supported by the recent analysis of a p53 mouse mutant with point mutations in the proline-rich domain (Toledo et al., 2007). As detailed below, this regulation model also has implications for the design of anticancer therapies based on the p53 pathway.
4. Medical Applications: the search for MDM2 and MDM4 antagonists
Inhibiting MDM2 activity in tumors that express wild-type p53 but have amplified the MDM2 gene has been considered an attractive anticancer strategy for many years (reviewed in (Vassilev, 2007). Different approaches have been used to find MDM2 antagonists. Analysis of the MDM2-p53 interface by crystallography indicated that only 3 N-terminal amino acids in p53 are essential for MDM2-p53 interactions (see above), suggesting that small peptides or molecules that efficiently disrupt this interaction could be found. A first series of experiments by David Lane’s group validated this approach, as p53 BoxI-derived peptides (called SuperTip) were found to specifically disrupt p53-MDM2 interactions. The efficiency of such peptides was reduced however, because they could form a helical structure when bound to MDM2, but adopted several conformations in solution. Different strategies were then designed to stabilize the helical structure of these peptides in solution. For example, hydrocarbon stapling recently led to the design of a promising peptide, though its efficacy remains to be tested in vivo (Bernal et al., 2007). A similar approach has consisted in the screening for chemical molecules that mimic p53 BoxI peptides to disrupt p53-MDM2 interactions. The earliest compounds, such as chalcone derivatives, were rather inefficient, but several promising molecules have been described in the past 3 years (reviewed in Vassilev, 2007). Among them, Nutlin 3a was shown to efficiently disrupt MDM2-p53 interactions in vitro, and its administration to nude mice bearing human xenografts tumors induced tumor shrinkage but appeared to not induce toxicity in the nude mice. It will be important to determine in more detailed studies whether toxicity is elaborated in any tissues of normal mice since conditional p53 activation in numerous tissues that lack Mdm2 induces cell death or arrest (Francoz et al., 2006; Ringshausen et al., 2006). Another approach involved searching for molecules that inhibit the MDM2 ubiquitin ligase activity. Compounds such as HLI98 inhibit MDM2-dependent p53 degradation, but also exhibit p53-independent effects and were not tested in vivo. Finally, a cellular-based screen led to the identification of RITA, which was proposed to bind p53 in such a way to antagonize MDM2 binding, though this remains to be proven. RITA appears to efficiently inhibit the growth of xenograft tumors derived from p53-wild type cell lines in SCID mice (Issaeva et al., 2004).
Importantly, these studies focused on the search for MDM2 antagonists to reactivate p53 in tumors, but a screening for specific MDM4 antagonists remains to be reported. Yet, the finding that MDM4 is frequently overexpressed in tumors, as well as studies that defined MDM4 as an important regulator of p53 activity (Figure 2), recently suggested that MDM4 could also be an interesting target in anticancer strategies. Direct evidence for this was obtained with the p53ΔP mouse mutant, when its capacity to suppress oncogene-induced tumors was analyzed in various MDM2 and MDM4 genetic contexts (Toledo et al., 2006). p53ΔP suppressed oncogene-induced xenograft tumors very poorly, and reducing the gene dosage for either MDM2 or MDM4 had no significant effect. However, a combined decrease in MDM2 and MDM4 gene dosage, or the complete ablation of MDM4, dramatically increased tumor suppression by p53ΔP. This demonstrated the relevance of MDM4 as a therapeutic target, and also showed that MDM2 and MDM4 antagonists could cooperate to induce strong p53 activation in tumors (Toledo et al., 2006). Furthermore, despite the similarity of p53-MDM2 and p53-MDM4 interfaces (Figure 1), the Supertip BoxI peptides, as well as Nutlin 3a, were found to be poor antagonists of MDM4-p53 interactions (Hu et al., 2006; Laurie et al., 2006; Patton et al., 2006; Wade et al., 2006). However, the combined use of Nutlin3a with MDM4 small interfering RNAs also supports the conclusion that MDM2 and MDM4 antagonists should together lead to more potent activation of p53 in tumor cells (Hu et al., 2006; Patton et al., 2006; Wade et al., 2006).
Further studies are required to evaluate the feasability of these approaches in a clinical setting (Brummelkamp et al., 2006; Ringshausen et al., 2006). The recent estimate that MDM2 and MDM4 antagonists could be used in the treatment of 2-3 millions patients diagnosed with cancer each year (Toledo and Wahl, 2006), provides a strong incentive for the search for p53-based anticancer strategies. While the potential toxicity of such drugs to normal cells remains an important issue to resolve, perhaps the « addiction » of tumor cells to overproduced MDM2 or MDM4 will make cancer cells uniquely vulnerable to p53 activating agents. In support of this, increased ARF levels in tumor cells apparently account for their increased sensitivity to p53 activation in vivo (Ventura et al., 2007). In addition, perhaps low, non toxic doses of p53 activating agents can be combined with non-toxic concentrations of standard cytotoxic chemotherapeutic agents to produce favorable therapeutic indices. Clearly, the ability to obtain diverse p53-activating strategies opens up exciting opportunites for the development and implementation of new therapeutic strategies for a large population of cancer patients.
Acknowledgments
The authors apologize for the omission of several references due to space limitations. This work was supported by Grant #4046 from the Association pour la Recherche sur le Cancer (to F.T) and Grants #CA100845 and CA061449 from the National Cancer Institute of the National Institutes of Health (to G.M.W).
Abbreviations
- ASPP
apoptosis-stimulating protein of p53
- BD
binding domain
- CNS
central nervous system
- dpc
days post-coitum
- HAUSP
Herpes virus-associated ubiquitin-specific protease
- MDM
murine double minute
- NLS
nuclear localization signal
- NES
nuclear export signal
- RING
really interesting new gene
- RITA
reactivation of p53 and induction of tumor cell apoptosis
- SCID
severe combined immune deficiency
- TAD
transactivation domain
- ZD
Zinc finger domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernal F, Tyler AF, Korsmeyer SJ, et al. Reactivation of the p53 Tumor Suppressor Pathway by a Stapled p53 Peptide. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesten LS, Zadelaar SM, De Clercq S, et al. Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell Death Differ. 2006;13:2089–2098. doi: 10.1038/sj.cdd.4401973. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Fabius AW, Mullenders J, et al. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- Chene P. Inhibition of the p53-MDM2 interaction: targeting a protein-protein interface. Mol Cancer Res. 2004;2:20–28. [PubMed] [Google Scholar]
- Chipuk JE, Green DR. Dissecting p53-dependent apoptosis. Cell Death Differ. 2006;13:994–1002. doi: 10.1038/sj.cdd.4401908. [DOI] [PubMed] [Google Scholar]
- Francoz S, Froment P, Bogaerts S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci USA. 2006;103:3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Gilkes DM, Farooqi B, et al. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem. 2006;281:33030–33035. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- Knights CD, Catania J, Giovanni SD, et al. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol. 2006;173:533–544. doi: 10.1083/jcb.200512059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie NA, Donovan SL, Shih CS, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- LeCam L, Linares LK, Paul C, et al. E4F1 is an atypical ubiquitin E3-ligase that modulates p53 effector functions independent of degradation. Cell. 2006;127:775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- Maetens M, Doumont G, De Clercq S, et al. Distinct Roles of Mdm2 and Mdm4 in Red Cell Production. Blood. 2007;109:2630–2633. doi: 10.1182/blood-2006-03-013656. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Wolff S, Erster S, et al. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Francoz S, Maetens M, et al. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13:927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- Marine JCW, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- Patton JT, Mayo LD, Singhi AD, et al. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 2006;66:3169–3176. doi: 10.1158/0008-5472.CAN-05-3832. [DOI] [PubMed] [Google Scholar]
- Ringshausen I, O’Shea CC, Finch AJ, et al. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Ronai Z. Balancing Mdm2 - a Daxx-HAUSP matter. Nat Cell Biol. 2006;8:790–791. doi: 10.1038/ncb0806-790. [DOI] [PubMed] [Google Scholar]
- Sheng Y, Saridakis V, Sarkari F, et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, et al. Acetylation of the p53 DNA-Binding Domain Regulates Apoptosis Induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Qu LK, Zhang J, et al. Critical role for Daxx in regulating Mdm2. Nat Cell Biol. 2006a;8:855–862. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, et al. Tip60-Dependent Acetylation of p53 Modulates the Decision between Cell-Cycle Arrest and Apoptosis. Mol Cell. 2006b;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Toledo F, Krummel KA, Lee CJ, et al. A mouse p53 mutant lacking the proline rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell. 2006;9:273–285. doi: 10.1016/j.ccr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Toledo F, Lee CJ, Krummel KA, et al. Mouse mutants reveal that putative protein interaction sites in the p53 proline-rich domain are dispensable for tumor suppression. Mol Cell Biol. 2007;27:1425–1432. doi: 10.1128/MCB.00999-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Trigiante G, Lu X. ASPP and cancer. Nat Rev Cancer. 2006;6:217–226. doi: 10.1038/nrc1818. [DOI] [PubMed] [Google Scholar]
- Vassilev LT. MDM2 inhibitors for cancer therapy. Trends Mol Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Wade M, Wong ET, Tang M, et al. Hdmx modulates the outcome of p53 activation in human tumor cells. J Biol Chem. 2006;281:33036–33044. doi: 10.1074/jbc.M605405200. [DOI] [PubMed] [Google Scholar]
- Wallace M, Worrall E, Pettersson S, et al. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell. 2006;23:251–263. doi: 10.1016/j.molcel.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, et al. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci USA. 2006;103:3226–3231. doi: 10.1073/pnas.0508500103. [DOI] [PMC free article] [PubMed] [Google Scholar]