Abstract
Rationale and Objectives:
The clinical utility of interactive computer-aided diagnosis (ICAD) systems depends on clinical relevance and visual similarity between the queried breast lesions and the ICAD-selected reference regions. The objective of this study is to develop and test a new ICAD scheme that aims improve visual similarity of ICAD-selected reference regions.
Materials and Methods:
A large and diverse reference library involving 3000 regions of interests was established. For each queried breast mass lesion by the observer, the ICAD scheme segments the lesion, classifies its boundary spiculation level, and computes 14 image features representing the segmented lesion and its surrounding tissue background. A conditioned k-nearest neighbor algorithm is applied to select a set of the 25 most “similar” lesions from the reference library. After computing the mutual information between the queried lesion and each of these initially selected 25 lesions, the scheme displays the six reference lesions with the highest mutual information scores. To evaluate the automated selection process of the six “visually similar” lesions to the queried lesion, we conducted a two-alternative forced-choice observer preference study using 85 queried mass lesions. Two sets of reference lesions selected by one new automated ICAD scheme and the other previously reported scheme using a subjective rating method were randomly displayed on the left and right side of the queried lesion. Nine observers were asked to decide for each of the 85 queried lesions which one of the two reference sets was “more visually similar” to the queried lesion.
Results:
In classification of mass boundary spiculation levels, the overall agreement rate between the automated scheme and an observer is 58.8% (Kappa = 0.31). In observer preference study, the nine observers preferred on average the reference lesion sets selected by the automated scheme as being more visually similar than the set selected by the subjective rating approach in 53.2% of the queried lesions. The results were not significantly different for the two methods (p = 0.128).
Conclusion:
This study suggests that using the new automated ICAD scheme, the inter-observer variability related issues can thus be avoided. Furthermore, the new scheme maintains the similar performance level as the previous scheme using the subjective rating method that can select reference sets that are significantly more visually similar (p < 0.05) than when using traditional ICAD schemes in which the mass boundary spiculation levels are not accurately detected and quantified.
Keywords: Computer-aided diagnosis, Two-alternative forced-choice experiments, Mammography, Observer preference study, Template matching, Visual similarity
I. INTRODUCTION
Computer-aided detection (CAD) of breast abnormalities is rapidly becoming a well accepted clinical practice to assist radiologists interpreting screening mammograms [1, 2]. Studies have found that radiologists' attitude toward and acceptance of CAD-cued micro-calcification clusters and masses are substantially different [3, 4]. Due to the high sensitivity (e.g., > 98% [5]), a large number of radiologists heavily rely on CAD-cued results when searching for and identifying micro-calcification clusters depicted on mammograms [4], This practice substantially improves the efficiency of radiologists when interpreting screening mammograms and helps radiologists detect more subtle cancers associated with micro-calcifications [6]. However, the lower CAD sensitivity for mass detection and the higher false-positive rates reduce radiologists' confidence in CAD-cued masses [4]. As a result, although CAD schemes could potentially detect a larger fraction of false-negative cancers depicted as subtle masses [7, 8], radiologists frequently discard CAD-cued subtle masses in the clinical practice [9, 10]. A recent study reported that using current commercialized CAD systems did not increase cancer detection rate but significantly increase recall (false-positive) rate in the clinical practice [11]. To improve CAD performance and increase radiologists' confidence in CAD-cued masses, investigators have been developing interactive computer-aided diagnosis (ICAD) schemes to identify visually similar and clinically relevant mass lesions [12-15]. Once a suspected mass lesion is queried by the observer, ICAD scheme segments the queried lesion and computes the likelihood of this region being associated with cancer based on comparisons with sets of “similar” lesions CAD-selected from a reference library. These “similar” lesions with verified outcome are displayed on the ICAD workstation and used as a “visual aid” to assist the radiologist in his/her decision making. Preliminary observer performance studies suggest that using an ICAD concept could improve radiologists' performance in classifying between malignant and benign masses [12] as well as increase their confidence in the CAD-cued results [16].
Although providing observers with a visual aid is a promising concept, to be effectively used the reference lesions selected by ICAD must be considered as “clinically relevant” and “visually similar” to the queried suspected lesion. Previous studies demonstrated that due to the substantial difference between computer vision and human vision, ICAD-selected “similar” lesions are often not considered by observers as “visually similar” [13, 14]. As a result, when observers believe that ICAD scheme selected a “poor” set of reference images for comparison, they are likely to ignore this visual aid.
Our previous study has demonstrated that after the differences in a number of image features (i.e., region size and circularity) have been controlled, the poor visual similarity of the ICAD-selected reference lesions was mainly caused by the substantial difference of the lesion boundary spiculation levels between the selected lesions. Without an accurate and robust method to detect spiculated rays around the mass boundary and quantify mass spiculation levels, automated selection of visually similar mass lesions remains one of the most difficult technical challenges in ICAD development. In an attempt to improve the selection of the reference lesions with respect to visual similarity, we previously tested a subjective (interactive) rating method in an attempt to improve the visual similarity of the ICAD-selected reference mass lesions [14]. The boundary spiculation levels of all 3000 lesions stored in our reference library were rated by an experienced observer and the ICAD scheme was restricted in searching for similar reference lesions that were rated as having spiculation levels similar to the queried lesion. In a two alternative forced choice observer preference experiment involving nine observers, there was an overall significant preference (p<0.01) for the reference lesion sets selected by the interactive rating method in which the spiculation levels of all reference lesions and the queried lesion were rated by the same observer as compared with the lesions sets selected by a traditional non-interactive ICAD method [14].
Although the study suggested that in general human observers were quite sensitive to mass spiculation levels in determining “visual similarity” [14], our recent study highlighted the large inter-observer variability when rating mass spiculation levels. When three experienced radiologists independently rated a set of 240 randomly selected mass lesions using a nine category scale in order to divide the mass lesions into three groups that represented “none/minimal,” “moderate,” and “severe/significant” spiculation levels, there was a relatively low level of agreement among the three radiologists. Agreement rates between paired observers ranged from 41% to 59% (Kappa = 0.14 to 0.31) [17]. Because of the large inter-observer variability, when applying this subjectively interactive rating approach in the ICAD system, different observers may rate the same queried mass lesion into different spiculation level. As a result, ICAD-selected reference lesions using the subjective rating method may be considered “visually similar” by some observers and “not similar” by others (due to the disagreement in spiculation rating between these observers and the observers who pre-rated all reference mass lesions).
The visualized spiculation level of mass boundary has been well recognized as an important factor in classification between malignant and benign masses [18]. Several techniques have been developed and tested to detect and classify between spiculated and non-spiculated masses [19-21]. One group used the analysis of locally oriented edges (ALOE) and a binary decision tree (BDT) [19]. Since spiculation frequently appears as linear structures with a positive image contrast and the structures lie in all radial directions to the mass center, a second group used the gradient directions (orientation) at pixels on, or close to, spiculation to detect and classify spiculated masses [20]. A third group defined a 30-pixel-wide band around the segmented mass boundary contour and then used a threshold and labeling algorithm to detect and classify between spiculated and non-spiculated masses [21]. Despite these efforts, adequate detection of mass spiculation remains a technical challenge. In addition, without a “ground truth” it is often difficult to quantify and correctly classify masses into different severity categories of spiculation levels.
To overcome the difficulty of inter-observer variability and generate a consistent “match” between queried regions and ICAD-selected reference image sets, we developed a new automated scheme that detects spiculated rays and classifies masses into three groups of spiculation levels. The ICAD scheme then uses a two-step approach including conditioned k-nearest neighbor (KNN) algorithm and mutual information (MI) template matching to select the visually similar lesions. To test this new ICAD scheme, we conducted a two- alternative forced-choice observer preference study. It compared the “visual similarity” between queried mass lesion and two sets of selected “similar reference lesions,” one by the new ICAD scheme and the other by the previously reported scheme using a subjective rating method [14]. The goal of this study is to develop a new technique that automatically selects visually similar reference lesions in such a manner that the selected lesions are at least as well appropriated as those selected by our previously developed subjectively interactive rating method [14] used for comparison.
II. MATERIALS AND METHODS
A. Automated detection and classification of mass region spiculation levels
The new automated scheme for detecting and classifying mass boundary spiculation levels includes five steps. An example showing the detection results at each step is shown in Fig. 1. First, once a suspected mass is queried by an observer, the scheme searches for an initial growth seed that is defined as the pixel with local minimum digital value inside a window of 4mm × 4mm centered at the queried location. An adaptive multi-layer topographic region growth algorithm [22] is applied to define the initial mass boundary contour. Based on the result of the region growth, the scheme computes a set of 14 image features to represent the segmented lesion and its surrounding tissue background. The detailed definitions and computing methods for these 14 features have been previously reported [14]. A feature-based artificial neural network (ANN) is then applied to generate an initial detection score (the likelihood of the suspected region depicting an actual mass) [23]. The topographic region growth algorithm had been implemented and used in our previous CAD scheme [8]. It plays an important role in reducing false-positive detections in that it typically eliminates approximately 75%-80% of suspected lesions (i.e., from an average of over 15 to approximately 3 regions per image) identified by the Difference-of-Gaussian filtering method employed to identify all possible regions that may depict masses; thereby, sensitivity remains high (i.e., > 85% of image based sensitivity or > 95% case based sensitivity) [23]. In order to minimize the risk of over segmentation (penetration of the growth region into surrounding normal breast tissue), current growth threshold in each layer is conservatively controlled by local contrast measurement [22]. As a result, segmented mass lesions are typically slightly smaller than the actual masses depicted on the images (based on visual examination). In a fraction of mass lesions, the region growth algorithm can be “trapped” by local structures (e.g., a cyst) inside the masses resulting in partial segmentation. Fig. 1 (b) shows the segmented boundary contour of the mass shown in Fig. 1 (a) after applying the topographic region growth algorithm.
Figure 1.
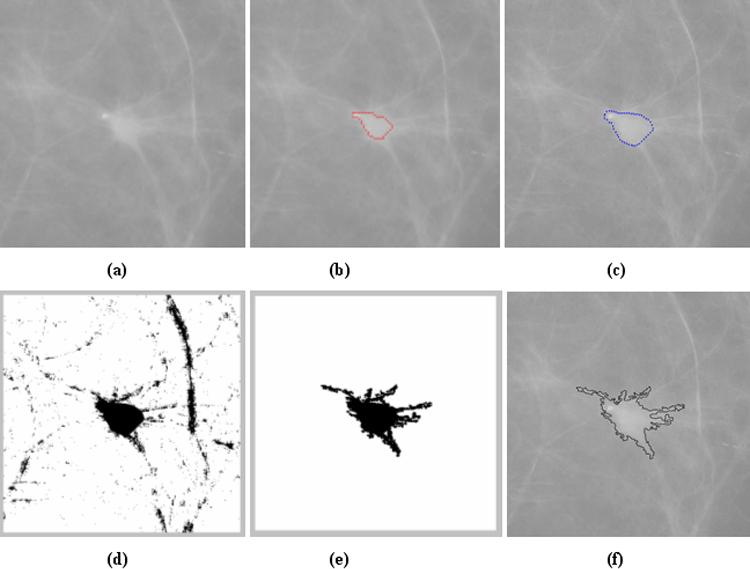
Example showing the five steps for detecting and classifying mass region spiculation. (a) ROI depicting a mass region. (b) Mass boundary resulting from the topographic region growth algorithm. (c) Mass boundary obtained from the active contour algorithm. (d) Initial detection of suspected pixels after a logical (“AND”) operation to combine the two maps of generalized gradient vector flow and positive contrast. (e) The final estimated mass region after the application of the morphological closing operation and labeling algorithm. (f) The computed perimeter pixels of the mass region.
Second, the scheme applies an active contour algorithm [24] to improve mass segmentation. The active contour is a deformable curve controlled by an internal and an external force. Selection of the initial growth boundary contour is important for avoiding the algorithm being trapped by local image noise [25]. In general, the internal force imposes a smoothness constraint on the contour, and the external force is typically determined by the magnitude of the image gradient and moves the vertices to locations with stronger gradients [26]. In mass segmentation, the assumption that the edge of a mass lesion always has the strongest gradients as compared with the surrounding background is frequently violated due to tissue overlap inherent to X-ray projection images. A fraction of subtle mass regions has fuzzy and ill-defined boundaries surrounded by dense and fluctuating tissue patterns. As a result, active contours can expand (penetrate) into the surrounding breast tissue. In our scheme, the boundary contour identified by the topographic region growth algorithm is used as the initial contour of the active contour algorithm and a map of generalized gradient vector flow representing the external forces [26] is computed. Unlike to what is the case of a typical active contour algorithm, shrinking is not allowed in our scheme in order to minimize the risk of being trapped inside the mass lesion. After completing a new active contour iteration, the scheme re-computes the same set of 14 features and applies the same ANN to compute a new detection score for the revised growth region. If the new detection score is higher than the previous one (indicating the higher likelihood of the new growth region being a true-positive mass region), the iteration of active contour continues. Otherwise, the iteration is terminated and the boundary contour generated by the previous iteration (with the higher detection score) is used as the final boundary contour. As shown in Fig. 1 (b) and Fig. 1 (c), after applying the active contour, the estimated mass boundary contour covers a larger area than when applying topographic region growth algorithm alone and it is substantially closer to the visually depicted boundary of the mass.
Third, the scheme detects linear structures (or initial suspected spiculated rays) surrounding the mass lesion segmented in step 2. For the first two steps, the region growth is conducted using sub-sampled images with pixel size of 400μm × 400μm to increase computation efficiency and reduce image noise. However, this step uses the original high resolution image with pixel size of 100μm × 100μm. The scheme typically extracts a 512 × 512 pixel region of interest (ROI) centered at the estimated center of the mass. The size of the ROI is large enough to cover the most breast masses of interest. For extremely large masses, ROI size is automatically increased by expanding the ROI 5mm away from the identified mass boundary in all four directions. The mass region boundary detected by the active contour is mapped from the low-resolution image to the high-resolution image. The scheme then computes two maps. The first one is a map of the generalized gradient vector flow [26] outside the mass as detected by active contour algorithm (step 2) and the second one is a “positive contrast” map. For the first map a pre-trained threshold value (e.g., 80 [17]) is applied to generate a binary representation of the gradient vector flow. The contrast of the pixel is defined for this purpose as the maximum pixel value difference between the center pixel (Ii) and any other pixel (In) inside a 1.5mm × 1.5mm convolution window (Ci = Max[In – Ii)], n = 1 to N). The scheme detects all pixels whose contrast values are larger than a pre-determined positive threshold value (e.g., = 150 in our scheme). Then, the scheme uses a logic “AND” operator to select all suspected pixels remained on both the gradient vector flow map and the positive contrast map. Other pixels (with lower contrast) detected in the gradient vector flow map are deleted [as shown in Figure 1(d)].
Fourth, the scheme applies a morphological closing operator to connect neighborhood (adjacent) regions that might be segmented separately during step 3 (i.e., the broken lines). A labeling algorithm is applied to all detected regions. Since all spiculation lines (or assigned pixels) must connect to the initial mass boundary identified in step 2, the scheme selects only one labeled region whose center is located inside the growth mass lesion and deletes all other labeled regions [as shown in Fig. 1 (e)].
Fifth, after connecting the detected “spiculated rays” to the mass lesion detected by the two region growth algorithms (steps 1 and 2), the scheme detects the perimeter pixels by applying a 3 × 3 window to convolve the image. If the window center overlaps with a pixel inside the mass and with at least one other pixel of the window located outside the mass lesion, this center pixel is defined as a “perimeter” pixel [as shown in Fig. 1 (f)]. The scheme then counts the total number of perimeter pixels (P) and the total number of pixels inside the mass (A). A “spiculation index” feature is computed to represent the spiculation level of the lesion. The larger the F value, the higher the estimated spiculation level is, since a larger number of pixels are classified as “perimeter” pixels.
B. Classification of three spiculation levels of reference lesions
We applied this new automated scheme to detect the boundaries of all 3000 suspected lesions (including 1000 malignant, 300 benign, and 1700 CAD-generated false-positives) contained in our reference library [14] and used the spiculation index to segment (classify) these mass lesions into three spiculation groups. The following approach was used to determine the two threshold values for this group segmentation. First, the spiculation levels of the 3000 reference lesions were grouped using the subjectively rated scores (1−9) as follows. All lesions with a subjective rating smaller than 4 were assigned to the first group (none/minimal spiculation), all lesions with the subjective rating between 4 and 6 (inclusive) were assigned to the second group (moderate spiculation), and all lesions with rating 7 and higher were assigned to the third group (severe/significant spiculation). The number of lesions in each group was counted and recorded. Then, the CAD-generated spiculation indices were sorted and two threshold values were determined to segment the 3000 lesions into three spiculation groups with respective number of lesions equal to those produced by the subjective, observer based rating method in each group.
C. A new scheme to select and display similar reference lesions
We developed and tested a new fully-automated ICAD scheme that searches in our reference library for mass lesions similar to an observer queried lesion. The scheme is composed of two steps. The first step uses a conditioned k-nearest neighbor (KNN) classifier to search for an initial set of “similar” lesions. The KNN classifier had been trained and optimized using genetic algorithm and ROC method as previously reported [14]. In brief, three boundary conditions on the difference in lesion size (area), circularity, and margin spiculation level between a queried lesion ( Aq, Cq and Sq ) and a reference lesion ( Ar, Cr and Sr ) are applied in the KNN classifier; they are: (1), (2) | Cr – Cq |≤ 0.15 , and (3) Sq = Sr . As a result, the KNN classifier is restricted to select “similar” lesions, each having a comparable size, an overall similar shape, and the same computed spiculation level to the queried lesion. Restricted by these three conditions, the KNN measures the “similarity” based on the difference of 14 feature values, fr (x) , between a queried ROI (yq) and a reference ROI (xi) in a multi-dimensional (n = 14) feature space:
The smaller the difference (“distance”), the higher the degree of the computed “similarity” between paired lesions (the queried lesion and the selected lesion). Computed distances between the queried lesion and each of the lesions in the reference library are recorded and sorted (rank ordered) from the smallest to the largest. The first K-nearest lesions in the rank ordered list are selected as the K “most similar” reference lesions. In our current study, K = 25.
The second step of the scheme uses a mutual information (MI) algorithm that aims to further improve visual similarity between the queried lesion and each of CAD-selected reference lesions. MI is originally defined as an intensity-based measure of general independence between two random variables x and y [27]. Applying MI concept to the two-dimensional images allows measuring the similarity between the pixel value distributions in two images. MI is widely considered as being one of the most effective approaches for the registration of multi-modality medical images [28] and for template matching of breast mass lesions [29]. To compute MI in our scheme, a rectangular window (ROI) is first drawn to cover the boundary of each selected lesion. Then, the size of the ROI is increased 6mm (e.g., 15 pixels in sub-sampled ROI with pixel size of 400μm × 400μm) in all four directions to include the surrounding normal tissue background used for image feature computation [14]. A same size ROI is mapped to each of the reference images. The MI of two compared ROIs, X (e.g., the queried ROI) and Y (e.g., the reference ROI), is computed as:
where, P(X,Y) is the joint probability density function (PDF) of the two ROIs, P(X) and P(Y) are the marginal PDFs. We used a histogram approach to compute the PDFs. In this approach, the joint PDF is estimated by computing the fraction of pixels in a particular pixel values bin in the 2-D histogram divided by the total number of pixels inside the ROI [30]. Before computing these PDFs, normalization of local histogram is applied to pre-process each paired ROIs in an attempt to reduce image noise and compensate the irregular variation or shift of the pixel value distributions. Specifically, the mean (μ) and the standard deviation (σ) of the pixel value distributions are calculated for each ROI. The interval [μ − 2σ, μ + 2σ] is divided into 128 pixel value bins. All pixels with values falling outside the interval range are assigned to the nearest ending bin during histogram calculation. After MI computation between the queried lesion and each of the reference lesions initially selected by the KNN classifier, the 25 “similar reference lesions” are re-sorted based on the computed MI values. The first N reference regions (e.g., N = 6) with the highest MI values in the sorted list are finally selected as the reference lesions “most similar” to the queried one. These lesions are displayed on an ICAD workstation for observers to visually compare between the queried lesion and the CAD-selected reference lesions. This aims at assisting radiologists in their decision making for diagnosis of the queried suspected mass lesion.
D. An observer preference study
To assess whether the new automated scheme improves visual similarity between the queried mass lesion and the CAD-selected reference lesions we performed a two-alternative forced-choice observer preference study. Since visual similarity is a subjective concept and there is no “ground truth,” absolutely objective similarity rating is difficult [12]. Hence, the two-alternative forced-choice observer preference study is considered a practical and effective approach for this purpose [31, 32]. The experimental design and data analysis procedure were similar to our previously reported study [14].
We selected 85 mass lesions from our reference library and defined these as “queried” lesions. For each queried lesion, two ICAD schemes were independently used to select two sets of six “most similar reference lesions” from our reference library. In the first scheme, the mass boundary spiculation levels of the queried and reference lesions had been subjectively rated by one experienced observer. In the second scheme, the new automated method is used to classify mass boundary spiculation levels. The two reference sets were displayed for comparison together with the queried lesion (Fig. 2) on our ICAD workstation. To better compare the difference in visual similarity in this study, we allowed for no more than one reference lesion to be the same in the two reference sets (Technically, the two methods might have selected the same reference lesion, or lesions, in some cases). A computer management program was implemented in the study to control the reading process from loading each queried mass, randomly displaying the two reference sets (left and/right to the queried lesion), and recording observer's preference.
Figure 2.
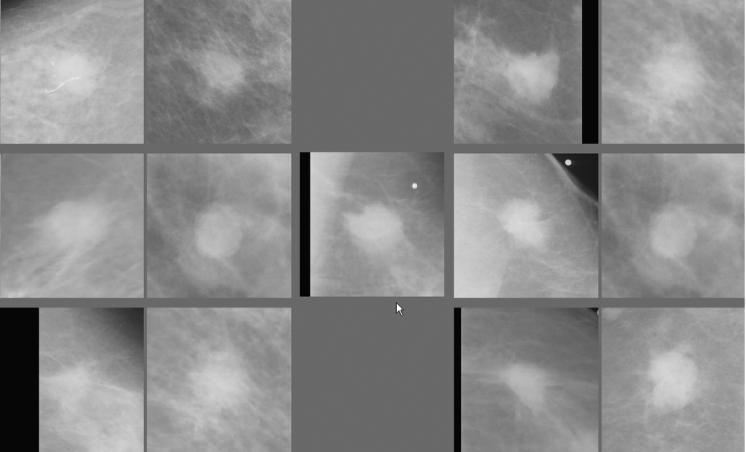
An example of a queried mass region (center as pointed by the arrow) and two sets of the reference regions selected by the subjective rating method (left) and the automated method (right).
Nine observers (including five board certified radiologists experienced in mammography and four investigators highly familiar with CAD research in mammography) participated in this observer preference study. Each observer visually examined the images displayed on the screen of the ICAD workstation and was forced to select one set of references (left group or right group) as being overall “visually more similar” to the queried lesion displayed in the center of the workstation screen (Fig. 2). We emphasized to each observer that he/she should make the choice based on the “overall” preference for one set (a group of six reference lesions) and not based on the “similarity” of any individual reference lesion in the set to the queried mass lesion. After reviewing a case, the observer selected one of the two sets by clicking the left button of the computer mouse with an arrow (mark) positioned anywhere inside the preferred set (left or right group). The preference selection was recorded and the next queried mass along with two new sets of selected reference lesions was instantaneously displayed for review and selection.
Recorded preference data were tabulated and compared for each individual observer and the group of nine observers (the average of preference results). One sample test for a binomial proportion (normal theory method with correction for continuity [33]) was used to test whether there was any significant difference in observers' preference for one of the two kinds of sets. A two sided test (p<0.05) was used for determining significance.
III. RESULTS
The agreement for rating boundary spiculation levels of mass lesions between the automated scheme and the subjective rating of one observer who rated spiculation levels of all reference lesions in the library is summarized in table 1. Among a total of 3000 reference lesions in the library, 1763 reference lesions were assigned to the same spiculation group (i.e., none/minimal, moderate, and severe/significant spiculation level) by two methods (subjective rating and the automated scheme) as shown in table 1. The overall agreement rate between two methods was 58.8% (with Kappa coefficient = 0.31).
Table 1.
Agreement between the number of reference regions classified into one of three spiculation categories as subjectively rated by an observer and the new automated scheme.
| Spiculation level | None/minimal | Moderate | Severe/significant | Total (Observer) |
|---|---|---|---|---|
| None/minimal | 499 | 410 | 9 | 918 |
| Moderate | 415 | 983 | 197 | 1595 |
| Severe/significant | 4 | 202 | 281 | 487 |
| Total (Scheme) | 918 | 1595 | 487 | 3000 |
Each of the nine observers completed the reading session in less than 30 minutes recording 85 forced choice preferences (selections) between the two sets of “similar” reference lesions presented for each of the queried mass lesion. The preferences of each of the observers are summarized in table 2. The five radiologists preferred between 45.9% and 63.5% of the reference image sets selected by the automated scheme as “more visually similar” to the queried masses of interest. The four non-clinician observers preferred the reference lesions selected by the automated scheme from 50.6% to 58.8% of the cases. The average preference level of the nine observers in identifying the reference sets of similar lesions selected by the automated method (53.2%) as “more similar” over the subjective rating method (46.8%) was not statistically significantly different (p = 0.128). Similarly, the differences were not significant for either of the observer groups (radiologists and non-radiologists).
Table 2.
Fraction (percent) of cases for which each observer preferred the reference set selected by the automated scheme as being "more visually similar" to the queried regions.
| Number of the Observer | 1 | 2 | 3 | 4 | 5 | Average |
|---|---|---|---|---|---|---|
| Radiologists | 63.5% (54 / 85) | 45.9% (39 / 85) | 54.1% (46 / 85) | 47.1% (40 / 85) | 51.8% (44 / 85) | 52.5% |
| Non-clinicians | 50.6% (43 / 85) | 50.6% (43 / 85) | 56.5% (48 / 85) | 58.8% (50 / 85) | N / A | 54.1% |
The number of cases for which a given number n of observers (n = 0 to 9) preferred the reference sets selected by the automated scheme as “more similar” is summarized in Fig. 3. This figure shows that for 9 of the 85 queried masses (10.6%), all nine observers preferred the reference image set selected by the automated scheme, while for 7 of the 85 queried lesions (8.2%), all observers preferred the reference set selected by subjective rating method. In 49 of the 85 queried cases (57.6%), more than half (at least 5 out of 9) of the observers preferred the reference sets selected by the automated scheme.
Figure 3.
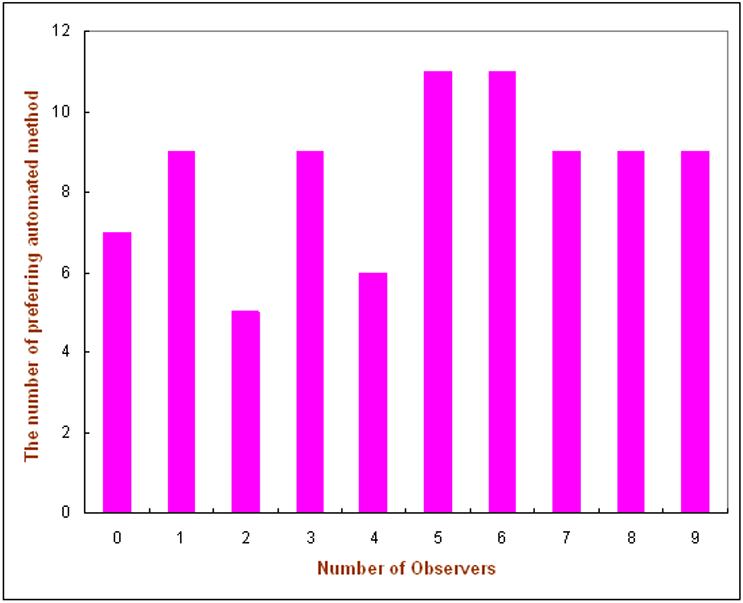
Number of cases (among the 85 ones) in which a given number of observers (n = 0 to 9) preferred the reference set selected by the automated scheme to the one selected by the subjective rating method.
IV. DISCUSSIONS
Our previous study demonstrated that the inclusion of a mass spiculation measure into the selection process for reference lesions could significantly improve the “visual similarity” between queried mass and ICAD-selected reference lesions [14]. However, visual rating of spiculation levels is highly subjective and somewhat ill-defined. Therefore, there is a large inter-observer variability when rating spiculation levels of mass region boundaries [17]. Different observers may rate the same queried lesion with different spiculation levels that may or may not match with the spiculation levels pre-rated and recorded by other observers in the reference library. As a result, the utility and performance of the ICAD system may be observer-dependent. ICAD scheme could not only select different reference lesions for different observers and might also generate different classification scores for the same queried suspect mass. To overcome these limitations (including inter-observer variability), we developed and tested a new automated scheme that aimed to consistently detect spiculation levels of masses and classify them into one of three groups (in the present study). The same scheme is used for both the queried lesions and each of the recorded lesions in the reference library.
Unlike previously developed computer schemes that detect and classify spiculated and nonspiculated masses [18-21], our scheme uses a simple summary index to quantify spiculation levels of any suspected masses. As do most of current CAD schemes, our scheme used the low-resolution image to define the initial boundary contour of a suspected mass, thereby reducing image noise and increasing the computation efficiency of the region growth algorithm. Our scheme then uses the high resolution image to detect spiculations connected to the lesion boundary and distributed along radial directions from the lesion center. By including the spiculated lines (if any), the scheme then computes a single spiculation related summary index (feature) to quantitatively classify mass spiculation levels. Although this feature, the square of perimeter divided by the mass area (), has been used in a number of CAD schemes [14, 34], the difference is that our automated scheme can detect more “spiculated rays.” Hence, the number of counted perimeter pixels is substantially increased for the spiculated masses. The agreement between the speculation levels assigned by this automated summary index and those assigned by the subjective rating of the single observer (Kappa = 0.31 as shown in table 1) is quite comparable to the agreement reported for the subjective ratings of two observers [17].
A number of different template matching approaches (or statistical models) have been investigated as the basis for similarity measures of breast masses [35]. Among these, information theory based mutual information (MI) is considered one of the most effective methods to search for similar mass regions depicted on different images [15]. However, the previously reported MI approach [29, 35] has two disadvantages related to the computing efficiency and measurement reliability when used in ICAD schemes. The different approach used in our study aimed at overcoming these disadvantages. First, using MI to search for similar regions requires significant computing time as the size of reference library increase. With our current ICAD workstation, however, the time for segmenting a queried lesion, computing the image related features, and identifying a set of “similar reference regions” from the library using the KNN classifier is less than one second because all features used by KNN have been pre-computed (off-line) and stored in the library. Using MI based measures to replace the KNN classifier, the process currently takes approximately 1.5 minute because all image processing routines of pixel value distributions must be conducted on-line. Although computing speed may be increased by code optimization and the use of a faster computer, the continually increasing size of the reference library in ICAD development rules out the use of a MI based approach as the primary one for real-time clinical applications. A previous study has also suggested that the success and efficiency of any method based on image content searching depend on how effective the system is in discarding the majority of irrelevant reference regions early in the process [36]. Therefore, to take advantage of the MI based approach in the measure of similarity we developed and tested a two stage scheme. The KNN feature based classifier quickly discards 99% of the 3000 reference lesions, and the MI related measure is only computed for a small subset of the “most similar” candidates (e.g., 25 in this study). Using our current ICAD workstation this two step process takes less than one second. Hence, when the observer interacts with our ICAD system, the “real-time” response can be achieved. Observers do not notice any time delay between querying a suspected lesion and viewing ICAD results.
Second, the previous studies applied MI to the ROIs with a fixed size (e.g., 512 × 512 pixels [29]). This approach reduces the reliability of MI results when comparing the masses with different sizes. In general, the accuracy of similarity measures of masses decreases as (1) the size of masses decreases and (2) the location of the masses is closer to the breast skin boundary. The sizes of ROIs used in our scheme for MI computation vary as the segmented sizes of the queried mass regions. Before computing MI, the majority of un-related normal (background) breast tissue are eliminated. Therefore, MI focuses on the comparison of segmented masses with controlled surrounding background tissue. The accuracy of MI-generated similarity measures is independent from the actual size of the queried mass lesions. This improves the reliability of MI-based similarity measures applied in ICAD schemes.
We are aware of the limitation of any measure of the agreement between spiculation levels as rated by an automated scheme and observers [17] as well as the shortcomings of using CAD performance (e.g., ROC or FROC) to evaluate the accuracy of visual similarity measures of ICAD schemes [15]. In the absence of “ground truth” the more relevant approach to evaluate the visual similarity of mass lesions selected by ICAD is an observer preference study [13, 14]. In this study we used the previously developed subjective rating method (that can select reference sets that are significantly more visually similar (p < 0.05) than when using the CAD schemes in which the mass boundary spiculation levels are not accurately detected and quantified [14]) as a new baseline for the comparison of the new automated scheme. We found that the performance of the automated scheme was comparable to that of the subjective rating approach, which suggests that this fully-automated scheme could replace the subjective rating method currently used in our ICAD system and generate consistent results in rating (or classifying) mass spiculation levels without reducing the visual similarity of selected reference lesions. Although a fraction of the 85 queried masses used in this study had already been used in our previous study [14], and despite the fact that seven of the nine observers had already participated in the previous observer preference study, we believe that the large time delay between the two studies (approximately 14 months), together with the random display of the queried lesions, significantly reduced (if not completely eliminated) any possible biases of the preference results recorded in this study.
Radiologists' confidence in and reliance on ICAD results for their decision making depend largely on whether they believe ICAD-selected reference lesions are clinically relevant and visually similar to the queried lesion. In this study, we only focused on the improvement of visual similarity between the queried masses and the ICAD-selected reference lesions with a fully-automated scheme. Because the reference library and the 14 image features used in this study were already used in our previous studies, the overall ICAD performance in classification between true-positive and false-positive (or malignant and benign) mass lesions should remain the similar level (e.g., the areas under ROC curves = 0.87) as we reported in two previously independent studies using the KNN classifiers with different learning methods and conditions [14, 37].
In summary, we have developed a new computer scheme to automatically detect spiculation in mass region and classify it in three groups. We then integrated the computed spiculation index into our new ICAD scheme that uses two stages (feature-based KNN classifier and MI based template matching) to select a set of reference lesions similar to the queried suspected mass. The results of the two-alternative forced-choice observer preference study demonstrate that reference region sets selected by this new ICAD scheme are comparable in “visual similarity” to those selected when using an subjective (interactive) rating method in which the queried lesion and all reference lesions were subjectively rated by the same observer. Therefore, the new ICAD scheme provides an alternative approach to consistently select reference lesions in real-time from a large reference library while maintaining high level of perceived visual similarity between selected reference sets and the queried mass. Although the results are encouraging, this is a very preliminary study. Further clinical and laboratory studies are needed to assess whether using this new ICAD scheme can significantly increase the cancer detection rate and/or reduce false-positive rate. The reading efficiency of radiologists when using ICAD also needs to be investigated in the future studies.
Acknowledgement:
This work is supported in part by Grants CA77850 and CA101733 to the University of Pittsburgh from the National Cancer Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Brem RF, Baum J, Lechner M. Improvement in sensitivity of screening mammography with computer-aided detection: a multi-institutional trial. Am. J. Roentgenol. 2003;181:687–693. doi: 10.2214/ajr.181.3.1810687. [DOI] [PubMed] [Google Scholar]
- 2.Taplin SH, Rutter CM, Lehman C. Testing the effect of computer-assisted detection on interpretive performance in screening mammography. Am. J. Roentgenol. 2006;187:1475–1482. doi: 10.2214/AJR.05.0940. [DOI] [PubMed] [Google Scholar]
- 3.D'Orsi CJ. Computer-aided detection: There is no free lunch. Radiology. 2001;221:585–586. doi: 10.1148/radiol.2213011476. [DOI] [PubMed] [Google Scholar]
- 4.Zheng B, Chough D, Cohen C, Sumkin JH, Abrams G, Ganott MA, Wallace L, Shah R, Gur D. Actual versus Intended Use of CAD Systems in the Clinical Environment. Proc SPIE. 2006;6146:9–14. [Google Scholar]
- 5.Brem RF, Hoffmeister JW, Zisman G, DeSimio MP, Rogers SK. A computer-aided detection system for evaluation of breast cancer by mammographic appearance and lesion size. Am. J. Roentgenol. 2005;184:893–896. doi: 10.2214/ajr.184.3.01840893. [DOI] [PubMed] [Google Scholar]
- 6.Freer TM, Ulissey MJ. Screening mammography with computer-aided detection: prospective study of 12,860 patients in a community breast center. Radiology. 2001;220:781–786. doi: 10.1148/radiol.2203001282. [DOI] [PubMed] [Google Scholar]
- 7.Warren Burhenne LJ, Wood SA, D'Orsi CJ, Feig SA, Kopans DB. Potential contribution of computer-aided detection to the sensitivity of screening mammography. Radiology. 2000;215:554–562. doi: 10.1148/radiology.215.2.r00ma15554. [DOI] [PubMed] [Google Scholar]
- 8.Gur D, Stalder JS, Hardesty LA, Zheng B, Sumkin JH. Computer-aided detection performance in mammographic examination of masses: assessment. Radiology. 2004;223:418–423. doi: 10.1148/radiol.2332040277. [DOI] [PubMed] [Google Scholar]
- 9.Khoo LA, Taylor P, Given-Wilson RM. Computer-aided detection in the United Kingdom National Breast Screening Programme: prospective study. Radiology. 2005;237:444–449. doi: 10.1148/radiol.2372041362. [DOI] [PubMed] [Google Scholar]
- 10.Ko JM, Nicholas MJ, Mendel JB, Slanetz PJ. Prospective assessment of computer-aided detection in interpretation of screening mammograms. Am. J. Roentgenol. 2006;187:1483–1491. doi: 10.2214/AJR.05.1582. [DOI] [PubMed] [Google Scholar]
- 11.Fenton JJ, Taplin SH, Carney PA, Cutter L, Sickles EA, D'Orsi C, Berns EA, Elmore G. Influence of computer-aided detection on performance of screening mammography. NEJM (New England Journal of Medicine) 2007;356:1399–1409. doi: 10.1056/NEJMoa066099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giger ML, Huo Z, Vyborny CJ, Lan L, Bonta I, Horsch K, Nishikawa RM, Rosenbourgh I. Intelligent CAD workstation for breast imaging using similarity to known lesions and multiple visual prompt aids. Proc SPIE. 2002;4684:768–773. [Google Scholar]
- 13.Muramatsu C, Li Q, Suzuki K, Schmidt RA, Shiraishi J, Newstead GM, Doi K. Investigation of psychophysical measures for evaluation of similar images for mammographic masses: preliminary results. Med Phys. 2005;32:2295–2304. doi: 10.1118/1.1944913. [DOI] [PubMed] [Google Scholar]
- 14.Zheng B, Lu A, Hardesty LA, Sumkin JH, Hakim CM, Ganott MA, Gur D. A method to improve visual similarity of breast masses for an interactive computer-aided diagnosis environment. Med Phys. 2006;33:111–117. doi: 10.1118/1.2143139. [DOI] [PubMed] [Google Scholar]
- 15.Tourassi GD, Harrawood B, Singh S, Lo JY, Floyd CE. Evaluation of information-theoretic similarity measures for content-based retrieval and detection of masses in mammograms. Med Phys. 2007;34:140–150. doi: 10.1118/1.2401667. [DOI] [PubMed] [Google Scholar]
- 16.Zheng B, Abrams G, Britton CA, Hakim CM, Lu A, Clearfield RJ, Drescher J, Maitz GS, Gur D. Evaluation of an interactive computer-aided diagnosis scheme for mammography: a pilot study. Proc SPIE. 2007:6515–42. [Google Scholar]
- 17.Zheng B, Abrams G, Leader JK, Park SC, Maitz GS, Gur D. Mass margins spiculations: agreement between ratings by observers and a computer scheme. Proc SPIE. 2007:6514–60. [Google Scholar]
- 18.Vyborny CJ, Doi T, O'Shaughnessy KF, Romsdahl HM, Schneider AC, Stein AA. Breast cancer: importance of spiculation in computer-aided detection. Radiology. 2000;215:703–707. doi: 10.1148/radiology.215.3.r00jn38703. [DOI] [PubMed] [Google Scholar]
- 19.Kegelmeyer WP, Pruneda JM, Bourland PD, Hillis A, Riggs MW, Nipper ML. Computer-aided mammographic screening for spiculated lesions. Radiology. 1994;191:331–337. doi: 10.1148/radiology.191.2.8153302. [DOI] [PubMed] [Google Scholar]
- 20.Karssemeijer N, Brake G. Detection of stellate distortions in mammograms. IEEE Trans Med Imaging. 1996;15:611–619. doi: 10.1109/42.538938. [DOI] [PubMed] [Google Scholar]
- 21.Sahiner B, Chan HP, Petrick N, Helvie MA, Hadjiiski LM. Improvement of mammographic mass characterization using spiculation measures and morphological features. Med Phys. 2001;28:1455–1465. doi: 10.1118/1.1381548. [DOI] [PubMed] [Google Scholar]
- 22.Zheng B, Chang YH, Gur D. Computerized detection of masses in digitized mammograms using single image segmentation and a multi-layer topographic feature analysis. Acad Radiol. 1995;2:959–966. doi: 10.1016/s1076-6332(05)80696-8. [DOI] [PubMed] [Google Scholar]
- 23.Zheng B, Sumkin JH, Good WF, Maitz GS, Chang YH, Gur D. Applying computer-assisted detection schemes to digitized mammograms after JPEG data compression. Acad Radiol. 2000;7:595–602. doi: 10.1016/s1076-6332(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 24.Yezzi A, Kichenassamy S, Kumar A, Olver P, Tannenbaum A. A geometric snake model for segmentation of medical imagery. IEEE Trans Med Imaging. 1997;16:199–209. doi: 10.1109/42.563665. [DOI] [PubMed] [Google Scholar]
- 25.Kupinski MA, Giger ML. Automated seeded lesions segmentation on digital mammograms. IEEE Trans. Med. Imaging. 1998;17:510–517. doi: 10.1109/42.730396. [DOI] [PubMed] [Google Scholar]
- 26.Xu C, Prince JL. Generalized gradient vector flow external forces for active contours. Signal Processing. 1998;71:131–139. [Google Scholar]
- 27.Cover TM, Thomas JA. Elements of Information Theory. Wiley; New York: 1991. [Google Scholar]
- 28.Pluim JP, Maintz JB, Viergever MA. Mutual-information-based registration of medical images, a survey. IEEE Trans Med Imaging. 2003;22:986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 29.Tourassi GD, Vargas-Voracek R, Catarious DM, Floyd CE. Computer-assisted detection of mammographic masses: a template matching scheme based on mutual information. Med Phys. 2003;30:2123–2130. doi: 10.1118/1.1589494. [DOI] [PubMed] [Google Scholar]
- 30.Maes F, Collignon A, Vandermeulen D, Marchal D, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16:187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 31.Paquerault S, Yarusso LM, Papaioannou J, Jiang Y, Nishikawa RM. Radial gradient-based segmentation of mammographic microcalcifications: observer evaluation and effect on CAD performance. Med Phys. 2004;31:2648–2657. doi: 10.1118/1.1767692. [DOI] [PubMed] [Google Scholar]
- 32.Balassy C, Prokop M, Weber M, Sailer J, Herold CJ, Schaefer-Prokop C. Flat-panel display (LCD) versus high-resolution gray-scale display (CRT) for chest radiography: an observer preference study. Am. J. Roentgenol. 2005;184:752–756. doi: 10.2214/ajr.184.3.01840752. [DOI] [PubMed] [Google Scholar]
- 33.Rosner B. Fundamentals of biostatistics. 5th edition Duxbury; Pacific Grove, CA: 2000. [Google Scholar]
- 34.Li L, Zheng Y, Zhang L, Clark RA. False-positive reduction in CAD mass detection using a competitive classification strategy. Med Phys. 2001;28:250–258. doi: 10.1118/1.1344203. [DOI] [PubMed] [Google Scholar]
- 35.Filev P, Hadjiiski L, Sahiner B, Chan HP, Helvie MA. Comparison of similarity measures for the task of template matching of masses on serial mammograms. Med Phys. 2005;32:515–529. doi: 10.1118/1.1851892. [DOI] [PubMed] [Google Scholar]
- 36.Huston L, Sukthankar R, Wickremesinghe R, Satyanarayanan M, Ganger GR. Diamond: a storage architecture for early discard in interactive search. Proc USENIX conference on File and Storage Technology; 2004; http://diamond.cs.cmu.edu/papers/fast2004-diamond.pdf. [Google Scholar]
- 37.Yang L, Jin R, Sukthankai R, Zheng B, Mummert L, Satyanarayanan M, Chen M, Jukic D. Learning distance metrics for interactive search-assisted diagnosis of mammograms. Proc SPIE. 2007:6514–52. [Google Scholar]


