Abstract
Genomics is now a core element in the effort to develop a vaccine against HIV-1. Thanks to unprecedented progress in high-throughput genotyping and sequencing, in knowledge about genetic variation in humans, and in evolutionary genomics, it is finally possible to systematically search the genome for common genetic variants that influence the human response to HIV-1. The identification of such variants would help to determine which aspects of the response to the virus are the most promising targets for intervention. However, a key obstacle to progress remains the scarcity of appropriate human cohorts available for genomic research.
Despite repeated exposures, some individuals do not seem to become infected with HIV-1, and among those that do, there is marked variation in how the virus is handled and in the time-course of progression to AIDS. It is known that host genetic differences contribute to this variation, but our knowledge of the relevant host genetic factors is currently limited for two main reasons: first, many studies have suffered from suboptimal study design, which is a common theme in the genetic-association literature1; second, a limited number of host genes have been studied so far, and these studies have focused heavily on candidate genes of acquired immunity and on proteins implicated in molecular studies of HIV-1 cell entry and proliferation2,3 (Table 1).
Table 1.
Grading evidence of candidate-gene studies of host susceptibility to HIV
| Gene | Function; role in HIV disease | Variants | Influence on HIV disease | Grading of genetic and molecular evidence* | ||||
|---|---|---|---|---|---|---|---|---|
| Effect size‡ | Amount and replication of evidence§ | Protection from bias∥ | Biological credibility¶ | Relevance** | ||||
| CCR5-CCR2 locus | Chemokine receptors; co-receptor of HIV-1 (CCR5) | CCR5 Δ32, CCR5 303T>A, CCR5 P1, CCR2 V64I, derived haplotypes | Protection (CCR5 Δ32, CCR5 303T>A, CCR2 V64I) or progression (CCR5 P1) | 1–3 | 3 | 3 | 3 | 3 |
| HLA | MHC; acquired immunity | HLA A, B, C homozygosity, or selected HLA B alleles | Protection or progression depending on allele | 1–3 | 3 | 3 | 3 | 3 |
| TRIM5α | Intrinsic immunity; HIV-1 restriction | Multiple non-synonymous variants | Neutral effect on HIV-1; effect on other retroviruses (N-MLV) | 0 | 2 | 2 | 3 | 1 |
| KIR | Innate immunity; regulation of NK cell response. | Specific KIR–HLA associations | Protection or progression depending on the specific association | 1 | 1 | 2 | 2 | 1 |
| CXCL12 (SDF-1) | Ligand of CXCR4 | 3′UTR SDF1-3′A | Neutral or progression | 1 | 2 | 2 | 1 | 1 |
| TSG101 | Vacuolar protein sorting; required for HIV-1 budding | Various haplotypes of promoter: −183T>C and intronic 181A>C | Protection or progression depending on haplotype | 1 | 2 | 2 | 1 | 1 |
| CCL5 (RANTES) | Ligand of CCR5 | Various haplotypes of promoter: −403G>A, −28C>G and intronic In1.1T>C | Protection or progression depending on haplotype | 1 | 1 | 2 | 2 | 1 |
| IL-10 | Anti-inflammatory cytokine | Promoter −592C>A | Progression | 1 | 1 | 2 | 2 | 1 |
| CCL3L1 (MIP1αP) | Ligand of CCR5 | Variable gene copy number | Progression associated with low-copy number | 1 | 1 | 2 | 2 | 1 |
| CX3CR1 | Fractalkine receptor; minor HIV-1 co-receptor | T280M | Progression | 1 | 1 | 1 | 2 | 1 |
| APOBEC3G | Intrinsic immunity; HIV-1 cDNA hypermutation | H186R or expression polymorphism | Progression | 1 | 1 | 2 | 1 | 1 |
| CCL3 (MIP1α) | Ligand of CCR5 | Intronic 459C>T | Progression | 1 | 1 | 2 | 1 | 1 |
| IL-4 | Pleiotropic cytokine | Promoter −589C>T | Conflicting | 0 | 1 | 1 | 2 | 1 |
Approximately 40 genes/genetic regions have been investigated for 10 years, under various study designs, populations and consensus3,78.
Proposed grading of genetic and molecular evidence: the degree of evidence is indicated from 1 (weak evidence) to 3 (strong evidence). The assessment reflects the interpretation of the authors of an unequal body of evidence. The grading of genetic and molecular evidence follows the proposal by J. P. Ioannidis1.
Effect size: 0, general consensus for no effect; 1, very small or small effect size (relative risk 2); 2, moderate effect size (relative risk 2–5); 3, large effect size (relative risk 5).
Amount and replication of evidence: 1, single or few scattered studies; 2, meta-analyses of group data; 3, large-scale evidence from inclusive networks.
Protection from bias: 1, clear presence of strong bias in the evidence; 2, uncertain about the presence of bias; 3, clear strong protection from bias.
Biological credibility: 1, no functional/biological data or negative data; 2, limited or controversial functional/biological data; 3, convincing functional/biological data.
Relevance: 1, no clinical or public health applicability; 2, limited clinical or public health applicability; 3, considerable clinical/public health applicability. Limited or insufficient data for: CXCR6, CCL4L, CCL2 (MCP1) CCL7, CCL1, IFNG, LTA (TNFβ), IL4R, IL2R, IL10R, DEFB1, CD209 (DC-SIGN), DC-SIGNR, MBL2, PTPRC, SLC11A1 (NRAMP1), VDR, PPIA, PML, TNFRSF1A, βTRC, INI1, NAF1, HP68, YY1, AIP1/ALIX, TAP, CTLA4 and PRF1. MHC, major histocompatibility complex; NK, natural killer cells; N-MLV, N-murine leukaemia virus; UTR, untranslated region.
The candidate-gene approach is based on a priori knowledge of the role or potential role of a gene in HIV pathogenesis. On selection of the candidate gene, the corresponding genomic region can be genotyped at known polymorphic positions, or re-sequenced with the purpose of identifying unknown variants. Association analysis can address the individual contributions of any single nucleotide polymorphism (SNP), or of a series of linked SNPs represented by a haplotype, to a study phenotype (Box 1). Statistical analysis needs to take into account issues such as multiple testing that will lead to an increasing number of false-positive tests as the number of SNPs, alleles, study endpoints, phenotypes or subgroups increase. Selected candidate SNPs are then followed by dedicated genetic, functional and biological testing to establish causality.
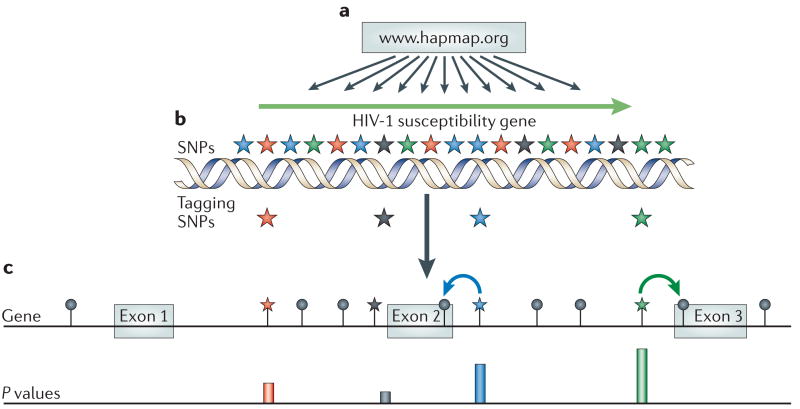
Box 1. The HapMap project.
The HapMap project (stands for Haplotype Mapping Project) provides an invaluable resource for researchers interested in relating human genetic variation to human health by allowing the selection of polymorphisms that represent other common polymorphisms in the human genome. These polymorphisms are called tagging single-nucleotide polymorphisms (tag SNPs) because they ‘tag’ other polymorphisms, which then do not need to be genotyped (see figure). The aim is to ensure that all the SNPs in the genome are directly represented by at least one SNP that is included on the analysis chip. Increasingly, this is being done on a genome-wide scale using standardized sets of tagging SNPs. This aim has nearly been achieved in current commercially available platforms for whole-genome association studies, allowing interrogation of most common human polymorphisms either directly or indirectly through association with tag-SNP candidates. As shown in panel a of the figure, genotype data (for a gene or a region of interest) are freely available from the HapMap website. The associations among SNPs in the data are then assessed to select tag SNPs. In part b of the figure, SNPs (coloured stars), that associate closely with each other have the same colour. The overall set of SNPs can be represented by a subset of tag SNPs, depicted in the figure as one star of each colour. Part c of the figure shows subsequent genetic association analysis. The tag SNPs are genotyped in a population sample in which individuals vary for the trait of interest, for example, increased susceptibility to HIV-1. A tag SNP that correlates with resistance to infection indicates that one of the known or unknown SNPs with which it is associated (black lollipops) might influence this phenotype — in this case, the non-genotyped SNP might be responsible for the association of the blue and green tag SNPs with resistance to HIV-1, shown by the corresponding high P values. From the set of SNPs that were not genotyped as tags, those in exons 2 and 3 are good candidates (coloured arrows depict a high linkage disequilibrium). Text and figure modified with permission from Nature Ref. 13 (2005) Macmillan Publishers Ltd.
Despite important efforts by various research groups, the candidate-gene approach has yielded limited results so far (Table 1). Only a fraction of the observed variability in the course of HIV-1 infection is explained by current knowledge2,4. The reasons for the limited success of this otherwise intuitively attractive hypothesis-driven approach is manifold, and include a limited knowledge of the attributes of candidate genes, lack of statistical power to detect small effects, incomplete knowledge of the genetic variation of the region, the inherent complexity of establishing the functionality of particular alleles and, until recently, the absence of efficient genotyping technologies. However, an optimist might point out that although only a modest number of genes have been assessed, and incompletely at that, there have nevertheless been unambiguous discoveries of important gene variants. Notable among these are the highly protective CCR5 Δ32 allele and the crucial role of selected human leukocyte antigen (HLA) alleles in disease progression.
The impact of HIV-1 infection on human health, the rapidity of expansion of the global HIV-1 epidemic, and the limitations of the candidate-gene approach argue for an immediate adoption of genome-wide association approaches. Here, genetic analyses assess the whole genome, or large genomic regions, with the goal of identifying genes associated with susceptibility to HIV-1, even in the absence of a priori knowledge of the most important genes. In this Perspective article, we discuss the new opportunities in HIV-1 research that have arisen through the development of high-throughput systems for SNP genotyping using information from the International Haplotype Mapping Project (or HapMap), and the use of comparative genomics to identify regions that are involved in host–pathogen genetic conflicts. Success in HIV-1 host genomic studies will depend on several key steps, notably, the identification of appropriate study phenotypes and the availability of large human cohorts for genetic studies5. The potential to harness genomic information for vaccine design will rest on the nature of the genes and alleles identified from the expected plethora of information emanating from host genomic projects.
Genomic opportunities
The heritable nature of susceptibility to HIV-1 can be observed in the familial segregation of a phenotype of resistance to infection, in studies of infected twins, and at the cellular level6–9. A significant heritable component of a phenotype constitutes the basis for genome analysis. Whole-genome studies can use data from families or from independent individuals; analysis of families can track the transmission of susceptibility alleles through generations. This approach, known as linkage analysis, is widely used in the mapping of monogenic disorders, and uses a few hundred polymorphic markers throughout the genome to identify the chromosomal region involved (for example, in rare familial syndromes of susceptibility to infectious diseases)10. However, family approaches are not feasible in the field of HIV-1 because of the limited familial nature of HIV-1 exposure (except in the setting of mother to child transmission). It is also known that family based approaches have limited power to detect gene variants that influence complex traits11. Therefore, studies of susceptibility loci in HIV-1 infection rely on population-based cohorts, using a denser map of genetic markers (hundreds of thousands) to do association studies between markers (such as SNPs) and the phenotype of interest. Genome approaches can also use data from other species, in particular from primates that show differences in the control of HIV-1 or of other lentiviruses, especially the simian immunodeficiency virus (SIV). These approaches are discussed further below.
HapMap and genetic-association studies
Over the past few years there has been a remarkable increase in our knowledge of the variation in the human genome and, today, most common human SNPs are known. In a particularly important effort, the HapMap (Box 1) identified the minimum number of SNPs that are needed to represent common human diversity. For this, the HapMap characterized patterns of association among different gene variants — the patterns of linkage disequilibrium across the genome — to select a minimal set of variants that are sufficient to represent common human variation in the context of an association study. These selected SNPs are referred to as tagging (tag) SNPs12,13 (Box 1).
In parallel with this increased knowledge, genotyping technologies have advanced significantly and now allow sufficient throughput to accommodate genome-wide approaches14. There is now a set of commercial whole-genome arrays using tag SNPs that represent variation in the human genome in non-African populations15,16. This means that the association of common variation with HIV-1 susceptibility can now in principle be satisfactorily captured by using sample sizes that are large enough to detect the effect of gene variants of modest impact. But key constraints remain. For example, it is known that rare variants will not be well represented in tagging strategies17. In addition, and of particular concern in the HIV-1 host genetics field, the current platforms for whole-genome analysis are known to represent variation in Africa inadequately. This asymmetry is both unfair and a constraint in HIV-1 genomics research and will need to be addressed directly as a priority. Not only is HIV-1 infection extremely prevalent in many parts of Africa, it is probable that there might be gene variants in Africa relevant to HIV-1 that have allele frequencies that are different from other populations. This is because of general patterns of population differentiation and possibly also because of recent selection16. This means that some genetic variants might be easier to find in some African populations in comparison with non-African populations and vice versa.
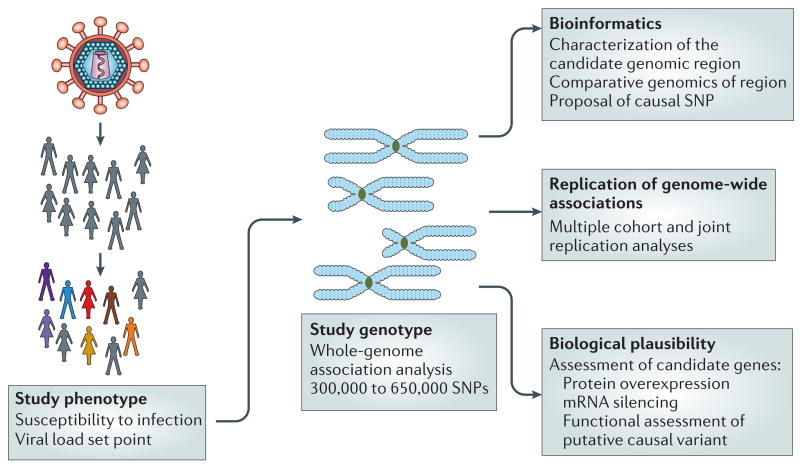
In Fig. 1, the design of a whole-genome association analysis of phenotypes with susceptibility to HIV-1 is shown, using a tagging strategy based on HapMap data to represent variation in the human genome. The design must take account of the known problems in the field. At the stage of study design, these problems include the undefined influence of the environment, and the difficulty of controlling for stratification, in particular linked to population or ethnic substructuring of sample sets, which is often also confounded by socioeconomic factors that influence infection and host response18. In addition, these studies present difficulties in defining and achieving adequate power and in establishing the computational and statistical analytical routines that are necessary to distinguish true- and false-positive discoveries and, more generally, to prioritize candidate genes to be investigated using genetic and functional approaches.
Figure 1. Whole-genome association analysis strategy.
The figure shows the steps needed to define a study phenotype and genotype, and the downstream procedures used to prioritize candidate genes to be investigated by genetic and functional approaches. The study phenotype(s) should be robust, that is, consistent, and should represent a characteristic of the individual that can be measured precisely. Examples include susceptibility to infection or viral load set point. Genotyping should take into consideration the specific population studied, as the number of polymorphic markers (single-nucleotide polymorphisms; SNPs) that require analysis differs for African or non-African populations. After the identification of genetic markers that are associated with the phenotype, a comprehensive bioinformatics and genetic analysis of the genomic region is done to identify causal candidate variants associated with the marker. Also, replication studies in other human cohorts, analysis in the context of other HIV-1 infection endpoints or phenotypes, and dedicated biological studies dictated by the nature of the genes that are mapped and their plausible mechanism of action are used to prioritize appropriate candidate genes.
Use of the HapMap resource
The availability of commercial products for whole-genome association analyses means that the use of the HapMap and other resources for genetic-association analyses has shifted from the upstream tasks of selecting what SNPs to genotype to the downstream tasks of interpreting the observed genotype–phenotype associations, and prioritizing the associations on the basis of their likelihood of being real rather than chance associations. However, this transition has occurred more rapidly than most researchers seem to have expected, and there are few tools available that allow ready interpretation of the results of whole-genome association studies. Considerable development is now needed to establish a framework for post-association annotation that will allow convenient high-throughput assessment of all those SNPs that show suggestive association with target phenotypes. Because real causal variants are likely to often show associations of a magnitude similar to those that show association purely by chance (because of the many polymorphisms that have been assessed) the degree of association for each of the SNPs must be assessed relative to features such as proximity to exons, location of conserved stretches of sequence and genomic regions with predicted regulatory function. The HapMap project will be a key tool in interpreting association results by allowing researchers to assess which polymorphisms are associated with the study phenotype (Box 1).
Ge and Goldstein have recently developed one tool for this purpose (see the Duke Institute for Genome Sciences and Policy in Further information), that uses available databases to draw the predicted gene structure, including sites of alternative splicing, and puts all the SNPs and their associated significance values onto this gene structure. Patterns of linkage disequilibrium throughout the gene are also indicated using HapMap data. These gene diagrams can then be used to identify the associations that seem most likely to be biologically relevant and can also be used to determine which genomic regions need to be resequenced to identify the causal polymorphisms that might be responsible for the association.
Comparative genomics
Just as humans differ in how well they handle HIV-1, non-human primate species differ in their handling of related retroviruses19. These differences include: resistance to infection (for example, differences in the susceptibility of primate cells to various lentiviruses)20,21; the satisfactory control of viral replication when infected (for example, in most experimentally infected chimpanzees, HIV-1 replicates poorly)22; and the occurrence of infections that are characterized by high-level replication without the hallmarks of disease progression (for example, in sooty mangabey and African green monkeys)23,24. One key observation is that naturally infected African primates do not develop immunodeficiency and have lower levels of T-cell immune activation and activation-induced cell death than HIV-1-infected humans. Recently, Schindler et al. reported that differences in the accessory viral protein Nef among primate lentiviruses (HIV compared with SIV) could explain the patterns of immune activation25. Nef alleles of HIV-1 fail to downregulate the T-cell receptor CD3 from infected cells thereby maintaining the responsiveness of infected T cells to activation25, in contrast to SIVs that downregulate CD3. However, there are several apparent exceptions to this view, and resolution of the role of Nef in immune activation awaits further investigation26.
Just as comparing the genomes of the various lentiviruses might explain important differences in immune pathogenesis between humans and non-human primates, exploring differences in the host genome can provide crucial insights into defence against retroviruses. Evolutionary genomic approaches have been proposed as powerful tools to identify regions in host proteins that are relevant for host–pathogen interactions27. These methods identify signs of positive selection or negative selection in the genome. A recent report from Voight et al. presents a map of recent positive selection in the human genome that shows the tremendous shifts experienced by modern human populations in habitats, food sources and population densities28. These signatures of selection are likely to be valuable signposts for gene variants that might influence medically relevant traits, including the susceptibility to infectious diseases, clearly one of the key selection pressures in human history. The availability of the complete chimpanzee and Rhesus macaque monkey genomes, and the ease of cross-species sequence amplification owing to the high degree of sequence conservation among humans and non-human primates allows comparative host genome analyses.
The first applications of evolutionary genomics to the field of HIV-1 have shown remarkable success. Comparative analysis of the primate antiretroviral cellular defence genes that encode APOBEC3G and TRIM5α (discussed below) has revealed the powerful selective pressures that have emerged from a long-standing battle between retroviruses and their hosts29–31. These proteins belong to a newly described form of innate immunity, coined ‘intrinsic immunity’, that assures protection by providing an ‘always-on’ line of defence, generally through intracellular obstacles to the replication of pathogens32. This component of the immune system is a cornerstone of the resistance of mammals against several classes of retroelements and retroviruses32.
Primate APOBEC3B, 3C, 3F and 3G have antiretroviral activity associated with the hypermutation of viral DNA through cytidine deamination (for recent reviews see Refs 21,33). However, the best studied member in humans, APOBEC3G, fails to restrict HIV-1 owing to degradation induced by the HIV-1 accessory protein virion infectivity factor (Vif)34. By contrast, several primate APOBEC3G proteins show activity against HIV-1 (Refs 21,35,36). Analysis of APOBEC3G across primate species reveals many residues in the amino-terminal cytidine deaminase domain that are under positive selection, which coincides with the proposed region of interaction with Vif (Fig. 2). Analyses have also shown that amino-acid residue 128 is under positive selection, which fits with evidence that this amino acid discriminates among the various HIV and SIV Vif proteins35,36.
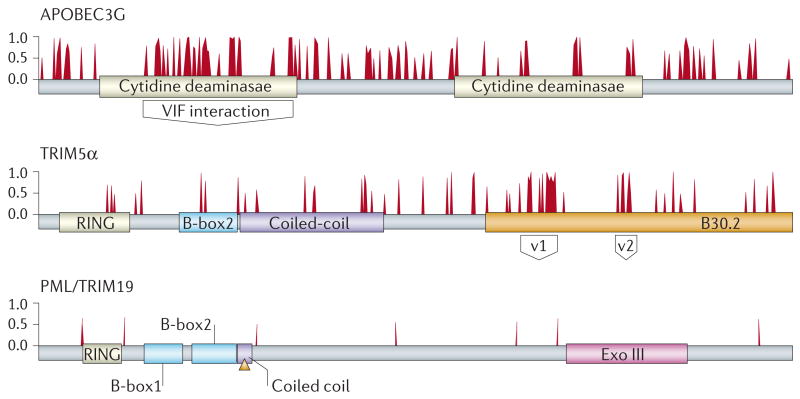
Figure 2. Comparative and evolutionary analysis of host proteins involved in HIV-1 pathogenesis.
This analysis used a comprehensive set of sequences that are representative of the primate lineages — new world monkeys, old world monkeys, gibbons and apes — to identify signs of positive and of negative selection in protein domains. The three proteins depicted represent different patterns of evolution in primates. The antiretroviral protein APOBEC3G is characterized by many residues that are under positive selective pressure, with a large cluster that delineates the virion infectivity factor (Vif)-interaction domain. These amino acids contribute to discrimination among the various Vif proteins of HIV and different SIVs. A second antiretroviral protein, TRIM5α (tripartite motif 5α) has a patch of positively selected residues that defines the variable 1 region (v1) and variable 2 region (v2) and that carry the capsid recognition specificity. The variable regions of TRIM5α might have evolved independently to recognize various retroviruses. By contrast, there are no residues under positive selective pressure in the PML/TRIM19 protein, which is also proposed to have antiviral properties. Therefore, the effect of PML/TRIM19 might be indirect. Failure to identify a signature of positive selection militates against a more direct role for this protein in antiviral defence, because it would be expected that prolonged contact with several pathogens over long evolutionary time periods would have resulted in signatures of positive selection indicative of a genetic conflict. Bars represent amino acids that are predicted to be under positive selection in a grading of posterior probability from 0 to 1. The various protein domains are indicated. Adapted with permission from Ref. 31.
The tripartite motif (TRIM) family is a large family of proteins that are characterized by a structure comprising a RING domain, one or two B-box domains and a predicted coiled-coil region37. In addition, TRIM proteins have specialized carboxy-terminal domains38,39. Some TRIM proteins display antiviral properties that target retroviruses in particular20. The best studied antiviral TRIM protein, TRIM5α, is a retroviral restriction factor that targets the early steps of cellular infection20; TRIM5α specifically recognizes the viral capsid and promotes its premature disassembly40. Human TRIM5α has limited efficacy against HIV-1, whereas some primate TRIM5α proteins can potently restrict HIV-1 (for reviews see Refs 38,39). Analysis of the TRIM5α protein across primate species pin-points a patch of amino acids that is under positive selective pressure at variable 1 region (v1) and variable 2 region (v2) (Fig. 2). Chimeras of human TRIM5α that carry the v1 patch from other primates, or selected mutants in the variable regions, can restrict HIV-1 (Refs 41–44). The variable regions of TRIM5α might have evolved independently to recognize various retroviruses44; however, in the absence of a crystal structure of the molecule, the exact molecular mechanisms that underlie the evolutionary changes remain undefined.
Comparative and evolutionary genomic methods have been applied to the understanding of other components of innate immunity. These include the analysis of the vertebrate Toll-like receptor proteins (an example of evolutionary conservation at multiple levels)45, the DC-SIGN (CD209) family of C-type lectins46,47 and the KIR genes that encode the main functional receptors of natural killer (NK) cells in humans, in whom the evolutionary forces driving the genesis of NK receptors and their HLA ligands represent a concerted response to pathogens48. Signals of evolutionary selection have also been searched for in 168 genes that are related to immune function49.
One should expect evolutionary analysis to become routine in the initial assessment of proteins that are involved in the pathogenesis of HIV-1 and SIV infection and of genes identified through whole-genome association analysis. The identification of signs of positive selection and of patches of genetic conflict should be a criterion for biological analysis. Therefore, there is great interest in promoting the sequencing of whole genomes from many more primates. Importantly, this effort should include our most distant relatives, the new world monkeys (such as tamarins), and prosimians (such as lemurs), to provide the needed out group and reference to estimate the evolutionary history of the proteins. However, as indicated in a recent review50, the flood of data and analytical methods in evolutionary and comparative genomics raise many new challenges, in particular the inherent multiple comparisons problem in searching the entire genome for specific regions that show evidence of selection. This creates a large ‘opportunity space’ for finding regions that show unusual patterns of variation in populations, or divergence among species, and the appropriate methodologies to correct for the number of possible tests that could be constructed are not obvious. Therefore, there is a need for the clear demonstration of the usefulness of a series of new dedicated statistical tests, more rigorous demonstration of evidence for natural selection and the inclusion of functional evidence for candidate loci50.
Phenotypes for genomic analysis
Most genetic studies so far have focused on endpoints, such as ‘time to AIDS or death’, that represent complex phenotypes resulting from many potential influences. Studies have also compared infected individuals with individuals that remain HIV-1 free despite repeated exposure; however, there is often limited accounting for the degree of exposure51. Least satisfactory, many studies have compared the frequencies of certain alleles in the infected population with the frequency in the general population, which is an insensitive measure of the enrichment or depletion of a genetic variant that is putatively associated with susceptibility to infection. Modest genetic influences on a disease will be hard to trace if masked by the influences of environment or concurrent disease. Therefore, there is a pressing need for study phenotypes that can be measured precisely, that are least influenced by external factors and that best reflect the basic mechanisms of pathogenesis.
The most useful insights will come from studies that focus on specific aspects of the response to HIV-1 that are sufficiently precise and narrow to guide our understanding of how the immune system controls or fails to control the virus. The overall pattern of viral replication following infection is one of an initial rapid increase to a peak level followed by the establishment of a viral load set point (at which the level of viraemia remains constant), which persists for an extended period, often many years (Fig. 3). Little is known about what determines peak viral load or exactly how it relates to disease progression in humans. In the simian HIV-1 infection model, there is a strong correlation between peak viral load and the extent of CD4+ T-cell depletion during acute infection52. Up to 80% of CD4+ T cells are infected at peak viraemia and the proportion of CD4+ T cells that are destroyed is correlated with the peak viral load52. The simple relationship between viral load and CD4+ T-cell depletion could allow prediction of the level of viral control that is required to prevent early immune damage. However, it is difficult to capture the unique window of peak viraemia in human cohorts as it requires a substantial effort in HIV-1 surveillance53.
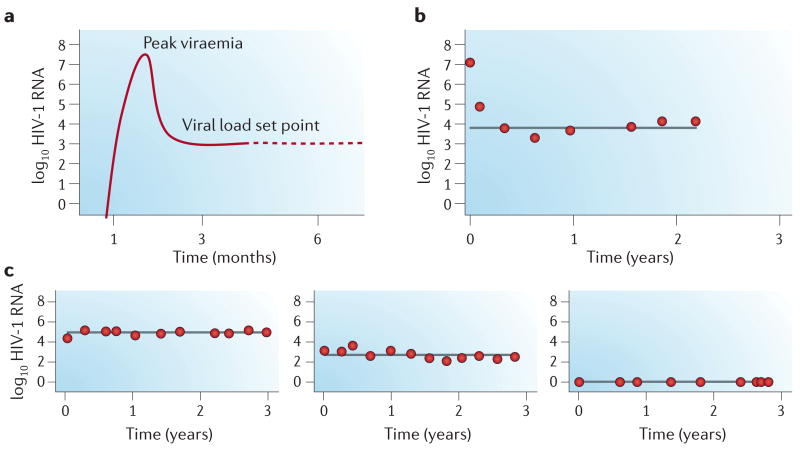
Figure 3. The viral load set point.
a | Soon after infection, HIV replicates to high levels (peak viraemia). Host factors then exert their effects to limit viral replication to a post-acute set-point level of plasma viraemia. b | The dynamics of establishment of the viral load set point in an individual presenting with HIV primary infection are shown. c | Viraemia data from three individuals are shown, characterized by post-acute viral load set points at 5, 3 and <2 log10 HIV-1 RNA copies per millilitre — fully stable over 3 years. Without these reliable patterns of inter-individual variation, genetics would have little to contribute to the analysis of viral load set point.
Similarly, the level of viraemia at set point is a determinant of the natural history of SIV infection54 and of the long-term prognosis of HIV-1 infection in humans55. The viral load set point is a particularly attractive target as a study phenotype. First, it is known that individuals vary by several orders of magnitude in the amount of virus per ml of blood at set point8, as illustrated in Fig. 3. For instance, there are rare individuals whose set point is at a level of virus that is essentially not detectable56. Second, the viral load set point is a characteristic of the individual. So far, there have been no large-scale genomic studies to determine the source of this variation. The identification of gene variants that are associated with the variation in viral load set point could implicate particular aspects of immune control. Peak and set-point viraemia are also of direct interest because they predict the degree of infectiousness of the individual57.
The fact that the viral load set point is established so early in the course of HIV-1 infection and that it seems to be particularly resistant to external variants over prolonged periods suggests that the environment might be only a small contributor. Therefore, the viral load set point seems to be determined mainly by two variables: influences from the host genome and the viral strain genome. Of course it cannot be ruled out that early environmental factors in an individual's life affect his or her immune system in such a way as to influence the set point that will be established on infection.
Another phenotype of significant interest for genetic studies is the status of exposed non-infected individuals. Indeed, the first identified gene variant that influenced susceptibility to HIV-1, CCR5 Δ32 was discovered using this framework. Several studies have examined heterosexual couples that are discordant for HIV-1 serostatus, female sex workers and men having sex with men who are highly exposed to HIV-1. The mechanisms identified or invoked to modulate susceptibility to infection in the various studies emphasize the relevance of differences in acquired immunity through the role of protective cytotoxic T-cell responses and NK activity in the context of specific HLA class I alleles, as well as differences in humoral responses at mucosal surfaces58,59. A large study is needed to compare uninfected individuals with known and quantifiable exposure to HIV-1 with a large cohort of infected individuals that are carefully matched demographically.
The importance of reliable study phenotypes is intimately linked to the creation of appropriate cohorts for genetic analysis. Several recommendations for optimal design of genetic-association studies in clinical trials and cohorts are presented elsewhere3,60–62. Examples of initiatives that have integrated genetic data for future clinical research are: the Adult AIDS Clinical Trials Group (AACTG) Protocol A5128 (Ref. 63) and the National Institute of Allergy and Infectious Diseases (NIAID)-sponsored GENOMICS protocol. Both protocols establish the conditions for storing DNA for studies that were not planned when informed consent was provided, and for future analyses.
The two-genome paradigm
The genome of HIV-1 co-evolves with that of the host. This is more pronounced for HIV-1 than for many other human pathogens because of the recognized capacity of retroviruses to mutate and thereby escape from the immune response and to adapt to the host environment. The need for continuous evolution for both the host and the pathogen is illustrated by the Red Queen principle.
At a population level, this host–viral interaction can be detected in several ways. For example, major histocompatibility complex (MHC)-restricted immune responses might shape viral genetic diversity over time because the immune selective pressure forces the emergence of viruses with escape mutations that result in infected cells that are no longer recognized by cytotoxic T lymphocytes64–67. However, certain host genotypes (for example, HLA-B*57) have reproducible associations with successful control of HIV-1 viral load68, implying that these genotypes exert selection pressure on diverse viral populations that are difficult to evade. The genome of HIV-1 can also be investigated using evolutionary genomics tools to identify signals of recent positive selection69,70. In the future, genomic approaches might need to simultaneously address the genome of both the host and the pathogen.
Although here we focus on the host genome, it is important to consider the role of the virus strain diversity and fitness as an equally important factor in HIV-1 susceptibility and pathogenesis71,72. The hypervariable nature of retroviruses and the simultaneous presence of quasispecies in any given patient make the goal of controlling for viral diversity a challenging one. Biological validation of newly identified host factors will, as a first step, require the use of a limited set of laboratory or clinical isolates and, increasingly, of data derived from studies using SIV.
Genomics and HIV-1 vaccine development
Genomics provides many new possibilities for vaccine research. First, previous unsuccessful vaccine trials can be investigated to identify genetic variants that influenced the magnitude of an evoked immune response and thereby highlight reasons for their overall failure. For example, the failed VAXGEN trial included 5,403 HIV-negative volunteers in a randomized, placebo-controlled trial of a recombinant glycoprotein 120 vaccine73. There were 368 subjects who acquired infection after vaccination; 6.7% in the vaccine arm of the trail and 7.0% in the placebo arm The course of HIV-1 infection was comparable between the two groups. However, the VAXGEN trial reported that titres of neutralizing antibodies in vaccine recipients varied considerably among participants, therefore constituting a potential target for genome analysis74. So, host genetic analyses allow revisiting of past vaccine trials for analysis of a new order of biological endpoints and basic questions. Whole-genome analyses can provide a unique description of how host genetic variation influences the early stages of HIV-1 infection, the exposed and uninfected state, the generation of anti-HIV-1 neutralizing antibodies and the breadth of cytotoxic T-lymphocyte responses. In addition, comparative and evolutionary genomics can complement the analysis by pointing to species-specific aspects of disease susceptibility, in particular through the standardized analysis of the mechanisms of innate immunity and intrinsic cellular defence.
Some of the challenges and opportunities discussed above are being pursued as part of the recently funded Center for HIV/AIDS Vaccine Immunology (CHAVI) (Box 2). The host genetics team at CHAVI is attempting a detailed genetic investigation of the control of the earlier phases of HIV infection, focusing both on viral load set point and, more ambitiously, on the control of viral build-up during the acute phase of infection. A key feature of the CHAVI programme is to use a common genetic platform across multiple cohorts allowing both replication of associations and a careful assessment of how gene variants function at different points during infection and subsequent viral dynamics. In addition to the CHAVI genomics project, another initiative, the HIV Elite Controller Study, will apply genomic techniques to the investigation of people infected with HIV-1 who have been able to maintain viral loads at or below the limits of detection. This collaborative study will address the key viral, host genetic and immunological contributions to this extraordinary outcome of infection. However, data analysis remains a significant problem: developing analytical routines that are able to deal with the unprecedented quantity of genomic information will pose a considerable challenge to the statistical genetics community which, until recently, has been only modestly involved in HIV-1 host genetics studies75.
Box 2. The CHAVI genome initiative.
The Center for HIV-AIDS Vaccine Immunology (CHAVI; see Further information) is a significant component of the Global HIV Vaccine Enterprise77. Based at Duke University in Durham, USA, it includes investigators from institutions across the globe. CHAVI has included genomics as a core project in the quest for a vaccine against HIV-1. The genome initiative includes the establishment of a series of cohorts with appropriate phenotypes. The first target phenotype is the viral load at set point in individuals with a known date of seroconversion. There is the potential to study between 1,000 and 2,000 qualifying patients across different cohorts. CHAVI will progressively focus on the study of at least 2,000 exposed individuals that will include infected and non-infected individuals from several clinical sites in Africa. This study aims to identify genetic determinants of protection from infection. Genotyping will be done using chips designed explicitly for whole-genome association studies. These chips allow genotyping of polymorphisms that represent common variation in the populations studied in the HapMap project: 550,000 single nucleotide polymorphisms (SNPs) for the study of subjects of European ancestry, and approximately 650,000 SNPs in subjects of African ancestry to reflect the lower level of linkage disequilibrium in Africa.
Conclusions
It is both a scientific and a social priority to apply modern and powerful genomic analyses to the study of HIV-1 infection and to aid the understanding of other important human pathogens, such as malaria and tuberculosis. The population geneticist Andrew G. Clark compared these new ‘discovery sciences’ to the voyage of the HMS Beagle “…setting sail to who knows where, amassing genome data on our hard drives and pawing through it to discover things that have not been seen before.”76
Paradoxically, as technical (large-scale genotyping) and analytical issues (genetic statistics) are progressively solved, the main challenge is posed by the quality of cohorts. Despite 2006 being the 25th anniversary of the first reported case of AIDS, appropriate cohorts remain surprisingly undeveloped — there are no large acute-infection cohorts and only moderately sized seroconversion cohorts (limiting replication of data), and an overall lack of preparedness for genetic work (ethical and legal clearance, and appropriate informed consent for genetic studies). We should emphasize that it is not technology, but cohorts, that constitute the key limiting factor today. We believe that the solution to this problem lies in multinational collaborations to establish and pool cohorts to synergize with efforts such as the Global HIV/AIDS enterprise that is being spearheaded by the NIH and the Bill and Melinda Gates Foundation.
Acknowledgments
We thank B. Haynes, K. Shianna, S. Antonarakis and J. Beckmann for helpful comments, M. Ortiz for assistance with Fig. 2, and B. Ledergerber (Swiss HIV Cohort) for data for Fig. 3. Funding for our work is provided by the Swiss National Science Foundation and by the Center for HIV-AIDS Vaccine Immunology and the National Institute of Health.
Glossary
- Comparative genomics
The study of relationships among the genomes of different species.
- Evolutionary genomics
The study of how changes in the content and organization of genomes have contributed to the diversity of life and the pressures that have shaped genomes. Comparative and evolutionary genomics identify signatures of selection that can often identify functional structures of sequence that are not otherwise easily annotated.
- Haplotype
Stands for haploid genotype — a collection of single nucleotide polymorphisms (SNPs) in one chromosome that tend to occur together (that is are linked) in individuals.
- Linkage analysis
Genes that lie close to each other on a chromosome tend to be inherited together. Markers on this segment of the chromosome can therefore be used for tracking the gene associated with a study phenotype.
- Linkage disequilibrium
The condition in which the haplotype frequencies in a population deviate from the values they would have if the allelic variants at each locus were combined at random. In this situation the allele present at one locus can be used to predict the allele present at another and both alleles need not be typed in an association study.
- Negative selection
Also known as purifying selection. Occurs when natural selection results in the removal of alleles that are deleterious. This can result in conserved gene sequences being maintained between species over long periods of evolutionary time. Evolutionary analyses identify signs of positive or negative selection by estimating the rates of substitutions leading to an amino-acid change (non-synonymous), versus substitutions that do not result in a change of the amino-acid residue (synonymous change).
- Population differentiation
If two populations are separated from each other then the allele frequencies at a locus in the two populations might differ because of random drift and/or differential selection in the two environments.
- Positive selection
This occurs when natural selection favours a single allele and consequently the frequency of an allele at a genetic locus increases until it is fixed in the entire population.
- Red Queen principle
The interaction between host and parasite leads to a constant evolutionary process of adaptation and counter-adaptation. In Lewis Carroll's Through the looking glass the Red Queen tells Alice that “It takes all the running you do to keep in the same place.”
- SNP
Pronounced ‘snip’. Stands for single nucleotide polymorphism which is a DNA sequence variation that arises when a single nucleotide (A, T, C or G) in the genome sequence differs among members of the species.
- Stratification
If a cohort includes individuals from different subgroups or populations then spurious association will be identified for any marker showing allele frequency differences between the (unknown) subgroups if the phenotype of interest has a different incidence between the subgroups. This phenomenon is called stratification and is thought to contribute to many false-positive findings in the complex-trait literature.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Contributor Information
Amalio Telenti, Amalio Telenti is at the Institute of Microbiology, University Hospital, University of Lausanne, 1011 Lausanne, Switzerland.
David B. Goldstein, David B. Goldstein is at the Institute for Genome Sciences and Policy, Center for Population Genomics and Pharmacogenetics, and the Center for HIV/AIDS Vaccine Immunology, Duke University, Durham, North Carolina 27708, USA
References
- 1.Ioannidis JP. Commentary: grading the credibility of molecular evidence for complex diseases. Int J Epidemiol. 2006;35:572–578. doi: 10.1093/ije/dyl003. [DOI] [PubMed] [Google Scholar]
- 2.O'Brien SJ, Nelson GW. Human genes that limit AIDS. Nature Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 3.Telenti A, Bleiber G. Host genetics of HIV-1 susceptibility. Future Virol. 2006;1:55–70. [Google Scholar]
- 4.Bleiber G, et al. Use of a combined ex vivo/in vivo population approach for screening of human genes involved in the human immunodeficiency virus type 1 life cycle for variants influencing disease progression. J Virol. 2005;79:12674–12680. doi: 10.1128/JVI.79.20.12674-12680.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Todd JA. Statistical false positive or true disease pathway? Nature Genet. 2006;38:731–733. doi: 10.1038/ng0706-731. [DOI] [PubMed] [Google Scholar]
- 6.Draenert R, et al. Constraints on HIV-1 evolution and immunodominance revealed in monozygotic adult twins infected with the same virus. J Exp Med. 2006;203:529–539. doi: 10.1084/jem.20052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plummer FA, Ball TB, Kimani J, Fowke KR. Resistance to HIV-1 infection among highly exposed sex workers in Nairobi: what mediates protection and why does it develop? Immunol Lett. 1999;66:27–34. doi: 10.1016/s0165-2478(98)00182-5. [DOI] [PubMed] [Google Scholar]
- 8.Chang J, et al. Twin studies demonstrate a host cell genetic effect on productive human immunodeficiency virus infection of human monocytes and macrophages in vitro. J Virol. 1996;70:7792–7803. doi: 10.1128/jvi.70.11.7792-7803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciuffi A, et al. Entry and transcription as key determinants of differences in CD4 T cell permissiveness to HIV-1 infection. J Virol. 2004;78:10747–10754. doi: 10.1128/JVI.78.19.10747-10754.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Picard C, Casanova JL, Abel L. Mendelian traits that confer predisposition or resistance to specific infections in humans. Curr Opin Immunol. 2006 doi: 10.1016/j.coi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 12.Altshuler D, et al. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein DB, Cavalleri GL. Genomics: understanding human diversity. Nature. 2005;437:1241–1242. doi: 10.1038/4371241a. [DOI] [PubMed] [Google Scholar]
- 14.Syvanen AC. Toward genome-wide SNP genotyping. Nature Genet. 2005;37:S5–S10. doi: 10.1038/ng1558. [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nature Genet. 2006;38:659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- 16.Pe'er I, et al. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nature Genet. 2006;38:663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- 17.Ahmadi KR, et al. A single-nucleotide polymorphism tagging set for human drug metabolism and transport. Nature Genet. 2005;37:84–89. doi: 10.1038/ng1488. [DOI] [PubMed] [Google Scholar]
- 18.Tsai HJ, et al. Comparison of three methods to estimate genetic ancestry and control for stratification in genetic association studies among admixed populations. Hum Genet. 2005;118:424–433. doi: 10.1007/s00439-005-0067-z. [DOI] [PubMed] [Google Scholar]
- 19.Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 20.Stremlau M, et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 21.Cullen BR. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J Virol. 2006;80:1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharp PM, Shaw GM, Hahn BH. Simian immunodeficiency virus infection of chimpanzees. J Virol. 2005;79:3891–3902. doi: 10.1128/JVI.79.7.3891-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch VM. What can natural infection of African monkeys with simian immunodeficiency virus tell us about the pathogenesis of AIDS? AIDS Rev. 2004;6:40–53. [PubMed] [Google Scholar]
- 24.Silvestri G. Naturally SIV-infected sooty mangabeys: are we closer to understanding why they do not develop AIDS? J Med Primatol. 2005;34:243–252. doi: 10.1111/j.1600-0684.2005.00122.x. [DOI] [PubMed] [Google Scholar]
- 25.Schindler M, et al. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell. 2006;125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 26.Foster JL, Garcia JV. HIV pathogenesis: Nef loses control. Cell. 2006;125:1034–1035. doi: 10.1016/j.cell.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z. The power of phylogenetic comparison in revealing protein function. Proc Natl Acad Sci USA. 2005;102:3179–3180. doi: 10.1073/pnas.0500371102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:e275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortiz M, Bleiber G, Martinez R, Kaessmann H, Telenti A. Patterns of evolution of host proteins involved in retroviral pathogenesis. Retrovirology. 2006;3:11. doi: 10.1186/1742-4690-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangeat B, Trono D. Lentiviral vectors and antiretroviral intrinsic immunity. Hum Gene Ther. 2005;16:913–920. doi: 10.1089/hum.2005.16.913. [DOI] [PubMed] [Google Scholar]
- 33.Yu XF. Innate cellular defenses of APOBEC3 cytidine deaminases and viral counter-defenses. Curr Opin HIV/AIDS. 2006;1:187–193. doi: 10.1097/01.COH.0000221590.03670.32. [DOI] [PubMed] [Google Scholar]
- 34.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 35.Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 36.Schrofelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc Natl Acad Sci USA. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nature Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 39.Towers GJ. Restriction of retroviruses by TRIM5α. Future Virol. 2006;1:71–78. [Google Scholar]
- 40.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30 2(SPRY) Domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Li X, Stremlau M, Lee M, Sodroski J. Removal of arginine 332 allows human TRIM5α to bind human immunodeficiency virus capsids and to restrict infection. J Virol. 2006;80:6738–6744. doi: 10.1128/JVI.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human TRIM5α leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 44.Ohkura S, Yap MW, Sheldon T, Stoye JP. All three variable regions of the TRIM5α B30.2 domain can contribute to the specificity of the retrovirus restriction. J Virol. 2006;80:8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roach JC, et al. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci USA. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bashirova AA, et al. Novel member of the CD209 (DC-SIGN) gene family in primates. J Virol. 2003;77:217–227. doi: 10.1128/JVI.77.1.217-227.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barreiro LB, et al. The heritage of pathogen pressures and ancient demography in the human innate-immunity CD209/CD209L region. Am J Hum Genet. 2005;77:869–886. doi: 10.1086/497613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook JG, et al. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walsh EC, et al. Searching for signals of evolutionary selection in 168 genes related to immune function. Hum Genet. 2006;119:92–102. doi: 10.1007/s00439-005-0090-0. [DOI] [PubMed] [Google Scholar]
- 50.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 51.Telenti A, Ioannidis JP. Susceptibility to HIV – disentangling host genetics and host behavior. J Infect Dis. 2006;193:4–6. doi: 10.1086/498535. [DOI] [PubMed] [Google Scholar]
- 52.Davenport MP, et al. Influence of peak viral load on the extent of CD4+ T-cell depletion in simian HIV infection. J Acquir Immune Defic Syndr. 2006;41:259–265. doi: 10.1097/01.qai.0000199232.31340.d3. [DOI] [PubMed] [Google Scholar]
- 53.Pilcher CD, et al. Detection of acute infections during HIV testing in North Carolina. N Engl J Med. 2005;352:1873–1883. doi: 10.1056/NEJMoa042291. [DOI] [PubMed] [Google Scholar]
- 54.Lifson JD, et al. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J Virol. 1997;71:9508–9514. doi: 10.1128/jvi.71.12.9508-9514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mellors JW, et al. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 56.Lambotte O, et al. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin Infect Dis. 2005;41:1053–1056. doi: 10.1086/433188. [DOI] [PubMed] [Google Scholar]
- 57.Cohen MS. Thomas Parran Award Lecture: transmission and prevention of transmission of HIV-1. Sex Transm Dis. 2006;33:338–341. doi: 10.1097/01.olq.0000218879.96236.9d. [DOI] [PubMed] [Google Scholar]
- 58.Kaslow RA, Dorak T, Tang JJ. Influence of host genetic variation on susceptibility to HIV type 1 infection. J Infect Dis. 2005;191:S68–S77. doi: 10.1086/425269. [DOI] [PubMed] [Google Scholar]
- 59.Mazzoli S, et al. HIV-specific mucosal and cellular immunity in HIV-seronegative partners of HIV-seropositive individuals. Nature Med. 1997;3:1250–1257. doi: 10.1038/nm1197-1250. [DOI] [PubMed] [Google Scholar]
- 60.Marzolini C, Kim RB, Telenti A. In: Pharmacogenetics of antiretroviral agents in AIDS Therapy. Dolin R, Masur H, Saag MS, editors. Churchill Livingstone; 2006. [Google Scholar]
- 61.Colhoun HM, McKeigue PM, Davey SG. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 62.Freimer NB, Sabatti C. Guidelines for association studies in human molecular genetics. Hum Mol Genet. 2005;14:2481–2483. doi: 10.1093/hmg/ddi251. [DOI] [PubMed] [Google Scholar]
- 63.Haas DW, et al. A multi-investigator/institutional DNA bank for AIDS-related human genetic studies: AACTG Protocol A5128. HIV Clin Trials. 2003;4:287–300. doi: 10.1310/MUQC-QXBC-8118-BPM5. [DOI] [PubMed] [Google Scholar]
- 64.Lieberman J. Defying death — HIV mutation to evade cytotoxic T lymphocytes. N Engl J Med. 2002;347:1203–1204. doi: 10.1056/NEJMcibr022067. [DOI] [PubMed] [Google Scholar]
- 65.Moore CB, et al. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 66.Telenti A, Beckmann JS, Mallal S. HLA and HIV: modeling adaptation to moving targets. Pharmacogenomics J. 2003;3:254–256. doi: 10.1038/sj.tpj.6500200. [DOI] [PubMed] [Google Scholar]
- 67.Trachtenberg E, et al. Advantage of rare HLA supertype in HIV disease progression. Nature Med. 2003;9:928–935. doi: 10.1038/nm893. [DOI] [PubMed] [Google Scholar]
- 68.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 69.Chen L, Perlina A, Lee CJ. Positive selection detection in 40,000 human immunodeficiency virus (HIV) type 1 sequences automatically identifies drug resistance and positive fitness mutations in HIV protease and reverse transcriptase. J Virol. 2004;78:3722–3732. doi: 10.1128/JVI.78.7.3722-3732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang W, Bielawski JP, Yang Z. Widespread adaptive evolution in the human immunodeficiency virus type 1 genome. J Mol Evol. 2003;57:212–221. doi: 10.1007/s00239-003-2467-9. [DOI] [PubMed] [Google Scholar]
- 71.van Opijnen OT, Berkhout B. The host environment drives HIV-1 fitness. Rev Med Virol. 2005;15:219–233. doi: 10.1002/rmv.472. [DOI] [PubMed] [Google Scholar]
- 72.Quinones-Mateu ME, Arts EJ. Virus fitness: concept, quantification, and application to HIV population dynamics. Curr Top Microbiol Immunol. 2006;299:83–140. doi: 10.1007/3-540-26397-7_4. [DOI] [PubMed] [Google Scholar]
- 73.Gilbert PB, et al. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis. 2005;191:666–677. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]
- 74.Montefiori DC, et al. Demographic factors that influence the neutralizing antibody response in recipients of recombinant HIV-1 gp120 vaccines. J Infect Dis. 2004;190:1962–1969. doi: 10.1086/425518. [DOI] [PubMed] [Google Scholar]
- 75.Evans DM, Cardon LR. Genome-wide association: a promising start to a long race. Trends Genet. 2006;22:350–354. doi: 10.1016/j.tig.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 76.Clark AG. Genomics of the evolutionary process. Trends Ecol Evol. 2006;21:316–321. doi: 10.1016/j.tree.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 77.Coordinating committee of the global HIV/AIDS vaccine enterprise strategy for developing an HIV vaccine. PLoS Med. 2005;2:e35. doi: 10.1371/journal.pmed.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Telenti A. Host polymorphism in post-entry steps of the HIV-1 life cycle and other genetic variants influencing HIV-1 pathogenesis. Curr Opin HIV/AIDS. 2006;1:232–240. doi: 10.1097/01.COH.0000221598.79906.12. [DOI] [PubMed] [Google Scholar]