Abstract
RPLP1 is one of acidic ribosomal phosphoproteins encoded by RPLP1 gene, which plays an important role in the elongation step of protein synthesis. The cDNA of RPLP1 was cloned successfully for the first time from the Giant Panda (Ailuropoda melanoleuca) using RT-PCR technology, which was also sequenced, analyzed preliminarily and expressed in E.coli. The cDNA fragment cloned is 449bp in size, containing an open reading frame of 344bp encoding 114 amino acids. Alignment analysis indicated that the nucleotide sequence and the deduced amino acid sequence are highly conserved to other five species studied, including Homo sapiens, Mus musculus, Rattus norvegicus, Bos Taurus and Sus scrofa. The homologies for nucleotide sequences of Giant Panda PPLP1 to that of these species are 92.4%, 89.8%, 89.0%, 91.3% and 87.5%, while the homologies for amino acid sequences are 96.5%, 94.7%, 95.6%, 96.5% and 88.6%. Topology prediction showed there are three Casein kinase II phosphorylation sites and two N-myristoylation sites in the RPLP1 protein of the Giant Panda (Ailuropoda melanoleuca). The RPLP1 gene was overexpressed in E. coli and the result indicated that RPLP1 fusion with the N-terminally His-tagged form gave rise to the accumulation of an expected 18kDa polypeptide, which was in accordance with the predicted protein and could also be used to purify the protein and study its function.
Keywords: cDNA cloning, overexpression, RPLP1, acidic ribosomal phosphoprotein P1, the Giant Panda (Ailuropoda melanoleuca)
1. Introduction
Ribosome, a compact ribonucleoprotein (RNP) grain that catalyzes protein synthesis, consists of 4 RNA species and approximately 80 structurally distinct proteins 1, 2. It can be dissociated into a small subunit and a large one whose shape and structure are irregular and asymmetric. The large ribosomal subunit has a distinct lateral protuberance called the stalk, which is an important and essential structure involved directly in the interaction of the elongation factors with ribosome during protein synthesis 3, 4. In Saccharomyces cerevisiae, the ribosomal stalk is made of five components, the 32kDa P0 and four 12kDa acidic proteins, P1alpha, P1beta, P2alpha and P2beta 5. These proteins are called A-proteins (acidic) or P-proteins (the phosphorylated eucaryotic A-proteins), which are generally present in multiple copies on the ribosome and have isoelectric points in the range of pH 3 to 5, in contrast to most ribosomal proteins which are single copy and basic 6.
There is only one type acidic protein in prokaryotes while two protein families are found in eukaryotes, phosphoproteins P1 and P2, which presumably have different roles and interact with each other participating in protein synthesis 7, 8. The acidic ribosomal phosphoprotein P1 (RPLP1) is internally located while RPLP2 is more external in the stalk and the interaction between RPLP1 and RPLP2 is necessary for forming the stalk structure 7, 9-13. RPLP1, also known as P1, RPP1, MGC5215, FLJ27448, is one of components constituting the large ribosomal subunit and is located in the cytoplasm. Together with RPLP0, RPLP2 and the conserved domain of 28S rRNA, it constitutes a major part of the GTPase-associated center in eukaryotic ribosomes 14, 15. In addition, it can attach to ubiquitin and assists the ubiquitin in regulating numerous important cellular processes including apoptosis, transcription, and the progression of cell cycle 16.
RPLP1 gene was cloned and studied from many animals including Homo sapiens 17-19, Mus musculus 20, Rattus norvegicus 21, 22, Gallus gallus 23, Danio rerio 24, Bos Taurus 25, Anopheles gambiae 26, Drosophila melanogaster 27, Bombyx mori 28 etc. However, RPLP1 gene from the Giant Panda (Ailuropoda melanoleuca) has not been reported yet.
The Giant Panda living on the arrow bamboo is a rare species currently found only in China and has been endangered due to climatic changes and past hunting activities. Its population originally extended throughout most of southern and eastern China, northern Myanmar and northern Vietnam. But now it is limited to the west and the north of Sichuan province, the south of Gansu province, eastern Tibet and the southwest of Shanxi province. It is one of declining wild lives in the world and studies on it are increasingly concerned by the world community. For many years, studies on the Giant Panda have been mainly concentrated on fields such as breeding and propagation, ecology, morphology, taxology, physiology and pathological biochemistry. Recently, researches on the Giant Panda have been undergoing a change from the level of cell (mainly the chromosome level) to molecule. Most studies have focused on genetic diversity, parentage and phylogenesis etc, while reports on functional gene are handful 29-34. It would further our understanding of this rare species by molecular studies of genes such as RPLP1.
This study was conducted using RT-PCR technique to amplify the cDNA sequence of RPLP1 gene from the total RNAs extracted from the skeleton muscle of the Giant Panda, and then analyzed the sequence characteristics of the protein encoded by the cDNA and compared it with those of human and other mammalian species reported. We also overexpressed it in E. coli using pET28a plasmids. The study provides scientific data for inquiring into the hereditary traits of the gene from Giant Panda and formulating the protective strategy for the Giant Panda.
2. Materials and methods
Materials and RNA isolation
Skeletal muscle was collected from a dead giant panda at the Wolong Conservation Center of the Giant Panda, Sichuan, China. The collected skeletal muscle was frozen in liquid nitrogen and then used for RNA isolation.
Total RNAs were isolated from about 400mg of muscle tissue using the Total Tissue/Cell RNA Extraction Kits (Waton Inc., Shanghai, China) according to the manufacturer's instructions. The total RNAs extracted were dissolved in DEPC (diethypyrocarbonate) water, and kept at -70°C.
Primers Design, RT-PCR, Cloning of RT-PCR Products and Sequencing
The PCR primers were designed by Primer Premier 5.0, basing on the mRNA sequence of RPLP1 from Homo sapiens (BC003369), Mus musculus (BC058685), Rattus norvegicus (BC058151), Sus scrofa (DQ629169) and Bos taurus (BC102695). The specific primers of RPLP1 are as follows: RPLP1-F: 5′-CTTCCGAGGAAGCTAAGGCCGCGTT-3′; RPLP1-R: 5′-AGAGGTTTAGTCAAAAAGACCAAAG-3′.
Total RNAs were synthesized into the first-stranded cDNAs using a reverse transcription kit with Oligo dT as the primers followed by PCR amplification according to the manufacturer's instructions (Promega). After amplification, PCR products were separated by electrophoresis in 1.5% agarose gel with 1× TAE buffer, stained with ethidium bromide and visualized under UV light. The expected fragments of PCR products were harvested and purified from gel using a DNA harvesting kit (Omega, China), and then ligated into a pUC18 vector at 22°C for 12 hours. The recombinant molecules were transformed into E. coli complete cells (JM109), and then spread on the LB-plate containing 50μg/mL ampicillin, 200mg/mL IPTG and 20mg/mL X-gal. Plasmid DNA was isolated and digested by PstI and ScaII to verify the insert size. Plasmid DNA was sequenced by Huada Zhongsheng Scientific Corporation (Beijing, China).
Construction of the Expression Vector and Overexpression of Recombinant RPLP1
PCR fragment corresponding to the RPLP1 polypeptide was amplified from the RPLP1 cDNA clone with the forward primer, 5'-TCT GGA TCC CTC GCC CGC ACC ATG -3' (BamHI) and reverse primer, 5'- CTC AAG CTT TTA GTC AAA AAG ACC AA-3' (HindIII), respectively. The PCR was performed at 94°C for 2 minutes; 35 cycles of 30s at 94°C, 45s at 55°C and 1 minute at 72°C; 7 minutes at 72°C. The amplified PCR product was cut and ligated into corresponding site of pET28a vector (Stratagen). The resulting construct was transformed into E.coli BL21(DE3) strain (Novagen) and used for the induction by adding IPTG (isopropyl-b-D-thiogalactopyranoside) at an OD600 of 0.6 and culturing further for 5 hours at 37°C, using the empty vector transformed BL21(DE3) as a control. The recombinant protein samples were induced after 1, 2 and hours, and then separated by SDS-PAGE (27) and stained with Coomassie Brilliant Blue dye.
Data analysis
The sequence data were analyzed by GenScan software (http://genes.mit.edu/GENSCAN.html). Homology research of the Giant Panda RPLP1 compared with the gene sequences of other species ware performed using Blast 2.1 (http://www.ncbi.nlm.nih.gov/blast/). ORF of the DNA sequence was searched using ORF finder software (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Protein structure of the RPLP1 sequence cloned was deduced using PredictProtein software (http://cubic.bioc.columbia.edu/predictprotein/).
3. Results and discussion
Analysis of the cDNA of RPLP1 from the Giant Panda
A cDNA fragment of 448bp was amplified from the Giant Panda with primers RPLP1-F and RPLP1-R (Fig. 1). Blast research showed that the cDNA sequence cloned shares a high homology to the RPLP1 from some mammals reported, including Homo sapiens, Mus musculus, Rattus norvegicus, Bos Taurus and Sus scrofa. On the basis of the high identity, we concluded that we had cloned the cDNA encoding the Giant Panda RPLP1 protein. The RPLP1 sequence was submitted to Genbank (accession number: EF631972), containing the 5'- untranslated sequence in size of 97bp and the 3'-untranslated region in size of 7bp. An ORF of 344bp encoding 114 amino acids was found in the cDNA (Fig. 2).
Fig 1.
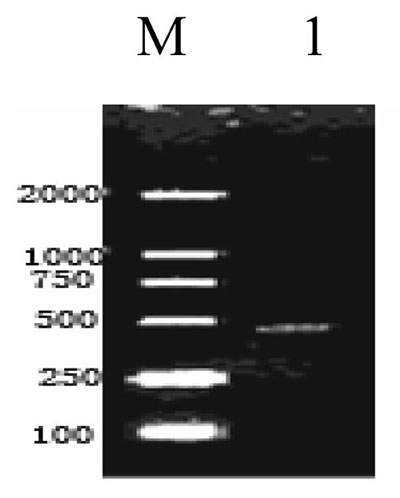
Reverse Transcription Polymerase Chain Reaction Products of the Giant Panda RPLP1. M: Molecular ladder DL2000; 1: the amplified RPLP1
Fig 2.
Nucleotide Sequence of cDNA Encoding the Giant Panda RPLP1 and the Amino Acid Sequence Deduced from Its ORF. Nucleotides are numbered in the 5'-to-3' direction. The predicted amino acid sequence of the gene is shown under the nucleotide.
Analysis of the homologies for nucleotide sequences and amino acid sequences
The homologies for nucleotide sequences between the Giant Panda and five mammals studied, including Homo sapiens, Mus musculus, Rattus norvegicus, Bos Taurus and Sus scrofa, are 92.4%, 89.8%, 89.0%, 91.3% and 87.5%, and for amino acid sequences are 96.5%, 94.7%, 95.6%, 96.5% and 88.6% (Table 1), respectively. Among them, the Giant Panda shares the highest homology with the nucleotide sequence and amino acid sequence of Homo sapiens. Alignment analysis of RPLP1 among the Giant Panda and the five mammals indicated that the nucleotide sequence and the deduced amino acid sequence are highly conserved (Table 1 and Fig. 3).
Table 1.
Comparison of nucleotide and protein sequence among 6 mammal species.
| A. melanoleuca | H.sapiens | M. musculus | R. norvegicus | B. taurus | S. scrofa | |
|---|---|---|---|---|---|---|
| A. melanoleuca | 92.4% | 89.8% | 89.0% | 91.3% | 87.5% | |
| H. sapiens | 96.5% | 89.6% | 89.0% | 93.3% | 88.7% | |
| M. musculus | 94.7% | 97.4% | 92.2% | 89.0% | 84.6% | |
| R. norvegicus | 95.6% | 98.2% | 97.4% | 89.0% | 86.4% | |
| B. taurus | 96.5% | 100.0% | 97.4% | 98.2% | 91.3% | |
| S. scrofa | 88.6% | 91.2% | 88.6% | 90.4% | 91.2% |
Note: The homology matrix of RPLP1 encoding sequence is above the diagonal, the homology matrix of protein sequence is below the diagonal.
Fig 3.
Comparison of the Amino Acid Sequence of RPLP1 Among the Giant Panda, Homo sapiens, Mus musculus, Rattus norvegicus and Bos Taurus. Sus: Sus scrofa; rat: Rattus norvegicus; bos: Bos Taurus; homo: Homo sapiens; mus: Mus musculus; panda: the Giant panda; __: casein kinase II phosphorylation site; ‗‗: N- myristoylation site
Prediction and analysis of protein functional sites in RPLP1 protein
Topology prediction revealed three N-glycosylation sites and two Casein kinase II phosphorylation sites in the RPLP1 protein. Our analysis indicated that there are three Casein kinase II phosphorylation sites and two N-myristoylation sites in the same locations in RPLP1 protein of the Giant Panda, Homo sapiens, Rattus norvegicus and Bos Taurus. Thus, in contrast with the topology prediction, there is one more N-myristoylation site in the RPLP1 from Mus musculus spanning amino acids 79-84 and one less N-myristoylation site in the RPLP1 from Sus scrofa spanning amino acids 57-62.
Further analysis detected 15 polymorphic sites in the amino acid sequences of the six species compared. Among these polymorphic sites, four amino acid residues in the RPLP1 of the Giant Panda are different from those of Homo sapiens and other four mammals. These distinctive sites are respectively I (Ile) at the fifty-fourth site, T (Thr) at the eighty-fifth site, D (Asp) at the hundredth site, H (His) at the one hundred-fifth site. All the four distinctive sites result from the transversion or transition of the corresponding codons, but there is not any deletion and insertion of bases. Among the four distinctive sites, the one at the hundred-fifth site is located in a casein kinase II phosphorylation site, but it does not result in any differences from human and other four mammalian species in the functional site. The fact shows that the variation of sites has no affect on the structure and function of RPLP1 protein. However, what changes caused by other mutations outside the functional sites in the structure and the function of RPLP1 protein are poorly understood and further studies need conducting.
It was reported that the carboxy-terminal 17-amino-acid sequence KEESEESD(D/E)DMGFGLFD of RPLP1 protein is the basis of its immunological cross-reactivity 17, 35. But for the RPLP1 protein from the Giant Panda, the 17-amino-acid sequence is KEDSEESHDDMGFGLFD. Within the area, there are two mutation sites, which have been underlined. One located at the one hundred-fifth site in a casein kinase II phosphorylation site changes from E to D, which is caused by its corresponding codon changing from GAT to CAT; the other one located at the hundredth site outside functional sites changes from D to H, which is caused by its corresponding codon changing from GA(A/G) to GAC. Further research needs to be conducted to explain whether the two mutation sites will lead to changes in the basis of its immunological cross-reactivity.
Prediction of the physical and chemical features of RPLP1 protein
The molecular weight of the RPLP1 protein from the Giant Panda is 11.566kDa and the theoretical pI is 4.40. Further analysis showed the molecular weight and theoretical pI of the putative protein among the Giant Panda and the five mammalians studied are very close (Table 2).
Table 2.
Molecular Weight and pI of RPLP1 Protein of the Giant Panda and Other Five Mammalian Species
| A. melanoleuca | H. sapiens | M. musculus | R. norvegicus | B. taurus | S. scrofa | |
|---|---|---|---|---|---|---|
| Molecular weight(kDa) | 11.566 | 11.5139 | 11.4749 | 11.4979 | 11.5139 | 11.7482 |
| pI | 4.40 | 4.26 | 4.28 | 4.28 | 4.26 | 4.26 |
Overexpression of the RPLP1 gene in E. coli
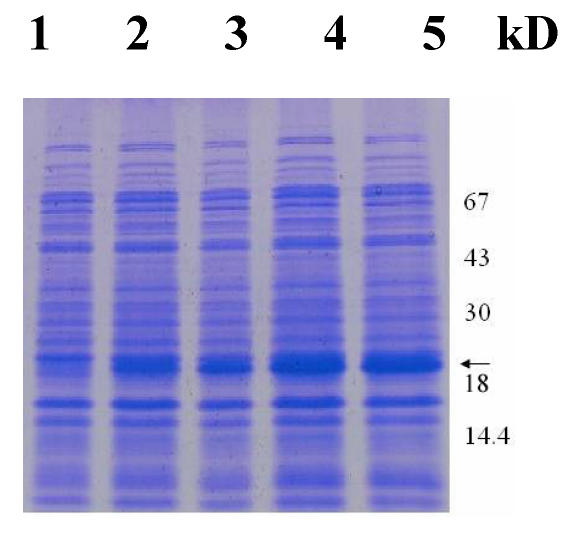
Next, we intended to overexpress the RPLP1 gene in E. coli using pET28a plasmids carrying strong promoter and terminator sequences derived from phage T7. For this purpose, the RPLP1 gene was amplified individually by PCR and cloned in a pET28a plasmid, resulting in a gene fusion coding for a protein bearing a His-tag extension at the N terminus. Expression was tested by SDS-PAGE analysis of protein extracts from recombinant E. coli strains BL21 (Fig. 4). Data showed that the protein RPLP1 fusion with the N-terminally His-tagged form gave rise to the accumulation of an expected 18 kDa polypeptide that formed inclusion bodies. Apparently, the recombinant protein was expressed after half an hour of induction and then after 2 hours reached the highest level. These results suggested that the protein is active and it is just the protein encoded by the RPLP1 from the Giant Panda. The expression product obtained could be used to purify the protein and study its function further.
Fig 4.
Protein extracts from recombinant E. coli strains were analyzed by SDS-PAGE gel stained with Coomassie blue. Numbers on right show the molecular weight, and the arrow indicates the recombinant protein bands induced by IPTG with 0, 0.5, 1, 2 and 4 hours (lane 1-5), respectively.
In Summary, the complete coding sequence of RPLP1 gene has been cloned using RT-PCR technology and the expression experiment in E.coli has been performed successfully. This is the first report on the RPLP1 gene from the Giant Panda. The data will not only enrich and supplement the information about RPLP1 but also allow the isolation of the structural gene from the Giant Panda. Further research on PRLP1 protein is desirable with a view of inquiring into the physiological function of the stalk structure. In addition, it will contribute to the protection for gene resources and the discussion of the genetic polymorphism of this “Treasure of China”, yet endangered species.
Acknowledgments
This work is supported by the Key Chinese National Natural Science Foundation (30470261), Application Technology Project in Sichuan Province (2006J13-057) and Key Discipline Construction Project in Sichuan Province (SZD0420).
References
- 1.Yoshihama M, Uechi T, Asakawa S et al. The human ribosomal protein genes: sequencing and comparative analysis of 73 genes. Genome Res. 2002;12(3):379–90. doi: 10.1101/gr.214202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang KC, Cui XS, Park SP et al. Identification of Differentially Regulated Genes in Bovine Blastocysts Using an Annealing Control Primer System. Mol Reprod Dev. 2004;69(1):43–51. doi: 10.1002/mrd.20156. [DOI] [PubMed] [Google Scholar]
- 3.Krokowski D, Bogueszewska A, Abramczyk D et al. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol Microbiol. 2006;60(2):386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Gbriel MA, Bou G, Briones E, Zambrano R et al. Structure and function of the stalk, a putative regulatory element of the yeast ribosome. Role of stalk protein phosphorylation. Folia Microbio (Praha) 1999;44(2):153–63. doi: 10.1007/BF02816234. [DOI] [PubMed] [Google Scholar]
- 5.Guarinos E, Remacha M, Ballesta JP. Asymmetric interactions between the acidic P1 and P2 proteins in the Saccharomyces cerevisiae ribosomal stalk. J Biol Chem. 2001;276(35):32474–9. doi: 10.1074/jbc.M103229200. [DOI] [PubMed] [Google Scholar]
- 6.Matheson AT, Moller W, Amons R Ribosomes: structure, functionand genetics. Baltimore: University Park Press; 1980. [Google Scholar]
- 7.Qiu D, Parada P, Marcos AG et al. Different roles of P1 and P2 Saccharomyces cerevisiae ribosomal stalk proteins revealed by cross-lingking. Mol Microbil. 2006;62(4):1191–202. doi: 10.1111/j.1365-2958.2006.05445.x. [DOI] [PubMed] [Google Scholar]
- 8.Remacha M, Jimenez-Diaz A, Santos C et al. Proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects. Biochem Cell Biol. 1995;73(11-12):959–68. doi: 10.1139/o95-103. [DOI] [PubMed] [Google Scholar]
- 9.Zurdo J, Parada P, van den Berg A et al. Assembly of Saccharomyces cerevisiae ribosomal stalk: bingding of P1 proteins is required for the interaction of P2 protiens. Biochemistry. 2000;39(30):8929–34. doi: 10.1021/bi000362j. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Gbriel MA, Remecha M, Ballesta JP. The RNA interaction domain but not the protein interacting domain is highly conserved in ribosomal protein P0. J Biol Chem. 2000;275(3):2130–6. doi: 10.1074/jbc.275.3.2130. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Fernandez J, Remecha M, Ballesta JP. The acidic protein binding site is partially hidden in the free Saccharomyces cerevisiae ribosomal stalk P0. Biochemistry. 2005;44(14):5532–40. doi: 10.1021/bi047332r. [DOI] [PubMed] [Google Scholar]
- 12.Ayub MJ, Barroso JA, Levin MJ et al. Preliminary structure studies of the hydrophobic ribosomal P0 protein from Trypanosoma cruzi, a part of the P0/P1/P2 complex. Protein Pept Lett. 2005;12(6):521–5. doi: 10.2174/0929866054395879. [DOI] [PubMed] [Google Scholar]
- 13.Tchorzewski M, Krokowski D, Rezeski W et al. The subcellular distribution of the human ribosomal “stalk” components: P1, P2 and P0 proteins. Int J Biochem Cell Bio. 2003;35(2):203–11. doi: 10.1016/s1357-2725(02)00133-4. [DOI] [PubMed] [Google Scholar]
- 14.Hagiya A, Naganuma T, Maki Y et al. A mode of assembly of P0, P1 and P2 proteins at the GTPase-associated center in animal ribosomal: in vitro analyses with P0 truncation mutants. J Biol Chem. 2005;280(47):39193–9. doi: 10.1074/jbc.M506050200. [DOI] [PubMed] [Google Scholar]
- 15.Remacha M, Jimenez-Diaz A, Santos C et al. Proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects. Biochem Cell Biol. 1995;73(11-12):959–68. doi: 10.1139/o95-103. [DOI] [PubMed] [Google Scholar]
- 16.Archibald JM, Evelyn M, Keelingl PJ. Novel ubiquitin fusion proteins: ribosomal protein P1 and actin. J Mol Biol. 2003;328(4):771–8. doi: 10.1016/s0022-2836(03)00374-7. [DOI] [PubMed] [Google Scholar]
- 17.Rich BE, Steitz JA. Human acidic ribosomal phosphoproteins P0, P1, and P2: analysis of cDNA clones, in vitro synthesis, and assembly. Mol Cell Biol. 1987;7(11):4065–74. doi: 10.1128/mcb.7.11.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gagou ME, Rodriguez Gabriel MA, Ballesta JP et al. Isolation and expression of the genes encoding the acidic ribosomal phosphoproteins P1 and P2 of the medfly Ceratitis capitata. Gene. 1999;226(2):365–73. doi: 10.1016/s0378-1119(98)00546-0. [DOI] [PubMed] [Google Scholar]
- 19.Shadeo A, Chari R, Vatcher G et al. Comprehensive serial analysis of gene expression of the cervical transcriptome. BMC Genomics. 2007;8:142. doi: 10.1186/1471-2164-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wool IG, Chan YL, Glück A et al. The primary structure of rat ribosomal proteins P0, P1, and P2 and a proposal for a uniform nomenclature for mammalian and yeast ribosomal proteins. Biochimie. 1991;73(7-8):861–70. doi: 10.1016/0300-9084(91)90127-m. [DOI] [PubMed] [Google Scholar]
- 21.Taulan M, Paquet F, Maubert C et al. Renal Toxicogenomic Response to Chronic Uranyl Nitrate Insult in Mice. Environ Health Perspect. 2004;112(16):1628–1635. doi: 10.1289/txg.7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wool IG, Chan YL, Glück A et al. The primary structure of rat ribosomal proteins P0, P1, and P2 and a proposal for a uniform nomenclature for mammalian and yeast ribosomal proteins. Biochimie. 1991;73(7-8):861–70. doi: 10.1016/0300-9084(91)90127-m. [DOI] [PubMed] [Google Scholar]
- 23.Ferro JA, Reinach FC. The complete sequence of a chicken-muscle cDNA encoding the acidic ribosomal protein P1. Eur J Biochem. 1988;177(3):513–6. doi: 10.1111/j.1432-1033.1988.tb14402.x. [DOI] [PubMed] [Google Scholar]
- 24.Uechi T, Nakajima Y, Nakao A et al. Ribosomal Protein Gene Knockdown Causes Developmental Defects in Zebrafish. PLoS ONE. 2006;1(1):e37. doi: 10.1371/journal.pone.0000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muramatsu Y, Lejukole HY, Taniguchi Y et al. Chromosomal assignments of expressed sequence tags for ACTG1, AHSG, COL1A1, GNAS1, and RPLP1 expressed abundantly in the bovine foetus. Anim Genet. 2002;33(3):230–1. doi: 10.1046/j.1365-2052.2002.t01-1-00876.x. [DOI] [PubMed] [Google Scholar]
- 26.Holt RA, Subramanian GM, Halpern A et al. The genome sequence of the malaria mosquito Ailuropod Anopheles gambiae. Science. 2002;298(5591):129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 27.Quesneville H, Bergman CM, Andrieu O et al. Combined evidence annotation of transposable elements in genome sequences. PLoS Comput Biol. 2005;1(2):166–75. doi: 10.1371/journal.pcbi.0010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouyanou S, Santos C, Koliaraki V et al. Protein BmP0 from the silkworm Bombyx mori can be assembled and is functional in the Saccharomyces cerevisiae ribosomal stalk in the absence of the acidic P1 and P2 proteins. Gene. 2003;314:173–9. doi: 10.1016/s0378-1119(03)00731-5. [DOI] [PubMed] [Google Scholar]
- 29.Montali RJ. Causes of neonatal mortality in giant panda. Tokyo Zoological Park Society. 1990:83–94. [Google Scholar]
- 30.Liao M J, Zhu M Y, Zhang Z H, Zhang A J. Cloning and sequence analysis of FSH and LH in the giant panda (Ailuropoda melanoleuca) Anim Reprod Sci. 2003;77:107–116. doi: 10.1016/s0378-4320(02)00275-0. [DOI] [PubMed] [Google Scholar]
- 31.Liao M J, Zhu M Y, Zhang Z H, Zhang A J. cDNA cloning of growth hormone from giant panda (Ailuropoda melanoleuca) and its expression in Escherichia coli. Comp Biochem Phys B. 2003;135:109–116. [PubMed] [Google Scholar]
- 32.Jennie P M, Alison M, Rong HL. Activins, inhibins, and follistatins: further thoughts on a growing family of regulator. Biol Med. 1992;201:1–15. doi: 10.3181/00379727-215-44130. [DOI] [PubMed] [Google Scholar]
- 33.Wu Z A, Liu W X, Murphy C et al. Satellite DNA sequence from genomic DNA of the giant panda. Nucleic Acids Res. 1990;18(4):1054. doi: 10.1093/nar/18.4.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou W, Chen Y, Peng Z et al. cDNA cloning and sequences analysis of ubiquinol-cytochrome c reductase complex ubiquinone-binding protein (QP-C) from giant panda. Acta Theriologica Sinica. 2007;27(2):190–194. [Google Scholar]
- 35.Lin A, Wittmann-Liebold B, McNally J et al. The primary structure of the acidic phosphoprotein P2 from rat liver 60S ribosomal subunits. J Biol Chem. 1982;257:9189–9197. [PubMed] [Google Scholar]