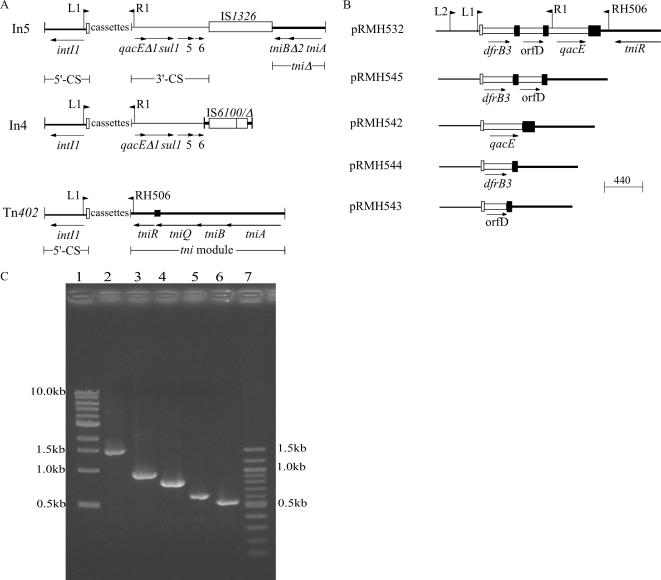
Amplification of the gene cassettes in class 1 integrons by PCR using primers in the 5′ conserved segment (5′-CS) and the 3′-CS (7, 8) has been used in hundreds of studies to identify integron-associated resistance genes (Fig. 1A). Equivalent PCR primers that detect cassettes in class 2 integrons, i.e., Tn7 family transposons (18), are also widely used. The approach used to detect integrons in antibiotic-resistant bacteria is to screen for the intI genes, using primers internal to these genes, and then amplify the cassettes in intI-positive strains by use of primers in the flanking conserved regions. Because different cassettes can have the same size and the arrays can include more than one gene (16), amplicon size alone cannot identify the cassettes, which are characterized by sequencing, PCR mapping (7), or restriction fragment polymorphisms (9, 10).
FIG. 1.
Context of gene cassettes in class 1 integrons. (A) Types of class 1 integron structure. Tall vertical bars represent the inverted repeats, and the various backbone segments, 5′-CS, 3′-CS, and the tni module, are represented by lines of different thicknesses. The short vertical bar represents the res site. Open vertical bars represent the attI site of class 1 integrons. Insertion sequences (IS) are shown as open boxes with the IS number above. Genes are indicated by arrows with gene names below. Positions of the primers used here are marked by flags with the primer name adjacent. L1 (5′-GGCATCCAAGCAGCAAGC-3′) and R1 (5′-AAGCAGACTTGACCTGAT-3′) (corresponding to primers 1 and 2 in reference 8) are representative of primer pairs used to amplify cassette arrays. (B) Cassettes in Tn402 and rearranged derivatives. The SphI-BamHI fragment of Tn402 present in pRMH532 is shown together with the cassette arrays present in the various derivatives recovered in this study. Cassettes are shown as open boxes with a vertical bar (representing the 59-be) at one end. (C) PCR amplification of gene cassettes using primer L1 with primer RH506. Lanes: 1, 1-kb markers; 2, pRMH532; 3, pRMH545; 4, pRMH542; 5, pRMH544; 6, pRMH543; 7, 100-bp markers. Predicted sizes of PCR products are 1,524 bp for pRMH532 and 924, 801, 617, and 534 bp for pRMH545, pRMH542, pRMH544, and pRMH543, respectively.
However, for some intI1-positive strains, a cassette PCR amplicon is not observed, and to date these strains have been largely ignored, even though such isolates can represent a significant proportion of the isolates studied (3). For class 1 integrons, if the sul1 gene found in the 3′-CS in both of the main structural types (12, 13) is not present, the absence of a PCR product may indicate that the priming site in the 3′-CS is missing. This can occur because the integron is recombinant with the 5′-CS of class 1 and the tns module of class 2 (14). Additionally, the 3′-CS is not found in Tn402 (15), the likely progenitor of class 1 integrons (12, 13). Instead, Tn402 includes a transposition gene module (tni module) containing tniA, tniB, tniQ, and tniR (also called tniC) (4, 5) (Fig. 1A). Several class 1 integrons with the tni module of Tn402 but a variety of gene cassettes have been reported (Table 1).
TABLE 1.
Cassette arrays flanked by the 5′-CS and tni modules
| Organism | Gene cassettes | GenBank accession no. | Reference |
|---|---|---|---|
| Klebsiella aerogenes | dfrB3-orfD-qacE | X72585 | 15 |
| Klebsiella pneumoniae | Unknown | U95134 | |
| Pseudomonas aeruginosa | blaVIM2-oxa2-aacA4-aadB-qacG | AY507153 | |
| Pseudomonas aeruginosa | aacA7-blaVIM2-dfrB5-aacC-A5 | AY943084DQ522233 | 11 |
| Uncultured | oxa2-aacA4-aadA1 | AJ744860 | 17 |
| Pseudomonas aeruginosa | blaVIM2-aacA4 | AM180753 | 6 |
| Pseudomonas aeruginosa | aacA7-blaVIM-2-dfrB5-aac6-II (ISPa21-like) | AM296017 |
To validate a method to detect cassettes in Tn402-type integrons, a fragment containing the cassette array of Tn402 from R751 was cloned into pACYC184, generating pRMH532 (Fig. 1B). IntI1-generated derivatives of pRMH352 that had lost one or more of the gene cassettes (Fig. 1B) were constructed as described previously (1, 2). Digestion with SphI and BamHI and sequencing were used to identify the cassettes and establish the mobility of the 587-bp qacE cassette, partial copy (bp 1 to 390) of which is in the standard 3′-CS. The cassette arrays were amplified using the 5′-CS primers L1 (5′-GGCATCCAAGCAGCAAGC-3′) (8) and L2 (5′-GACGATGCGTGGAGACC-3′) with primer RH506 (5′-TTCAGCCGCATAAATGGAG-3′) in the tniR gene (Fig. 1B). Plasmid DNA was prepared using a Wizard Plus SV Miniprep DNA purification kit (Promega). PCR amplification was carried out in PCR buffer (New England Biolabs) containing 160 μM of each deoxynucleotide triphosphate, 50 pmol of each primer, approximately 10 to 50 ng of template, and 1 unit of Taq DNA polymerase (Roche). Reaction conditions were 95°C for 3 min followed by 35 cycles of 95°C for 30 s, 64°C for 90 s, and 72°C for 90 s and by a final incubation at 72°C for 5 min. The sizes of the amplicons obtained with both L1 (Fig. 1C) and L2 and of RsaI restriction fragments were as predicted from the Tn402 sequence (GenBank accession no. X72585; Tn5090 is Tn402). This method should prove useful in analyzing strains that include intI1 but not sul1 and do not form an amplicon with the L1-R1 (or equivalent) primer pair.
Footnotes
Published ahead of print on 25 June 2007.
REFERENCES
- 1.Bunny, K. L., R. M. Hall, and H. W. Stokes. 1995. New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrob. Agents Chemother. 39:686-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collis, C. M., and R. M. Hall. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 174:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu, S.-C., T.-H. Chiu, J.-C. Pang, C.-H. Hsuan-Yuan, G.-N. Chang, and H.-Y. Tsen. 2006. Characterization of antimicrobial resistance patterns and class 1 integrons among Escherichia coli and Salmonella enterica serovar Choleraesuis strains isolated from humans and swine in Taiwan. Int. J. Antimicrob. Agents 27:383-391. [DOI] [PubMed] [Google Scholar]
- 4.Kholodii, G. Y. 1995. Inversion activity of the resolution of transposons Tn5053 and Tn402 possessing a nonstandardly organised res region. Genetika 31:1698-11703. [PubMed] [Google Scholar]
- 5.Kholodii, G. Y., S. Z. Mindlin, I. A. Bass, O. V. Yurieva, S. V. Minakhina, and V. G. Nikiforov. 1995. Four genes, two ends, and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol. Microbiol. 17:1189-1200. [DOI] [PubMed] [Google Scholar]
- 6.Lagatolla, C., E. Edalucci, L. Dolzani, M. L. Riccio, F. De Luca, F. Medessi, G. M. Rossolini, and E. A. Tonin. 2006. Molecular evolution of metallo-β-lactamase-producing Pseudomonas aeruginosa in a nosocomial setting of high-level endemicity. J. Clin. Microbiol. 44:2348-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lévesque, C., L. Piché, C. Larose, and P. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lévesque, C., and P. H. Roy. 1993. PCR analysis of integrons, p. 590-594. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. Mayo Foundation, Rochester, MN.
- 9.Levings, R. S., D. Lightfoot, L. D. H. Elbourne, S. P. Djordjevic, and R. M. Hall. 2006. New integron-associated gene cassette encoding a trimethoprim-resistant DfrB-type dihydrofolate reductase. Antimicrob. Agents Chemother. 50:2863-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levings, R. S., D. Lightfoot, S. R. Partridge, R. M. Hall, and S. P. Djordjevic. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lolans, K., A. M. Queenan, K. Bush, A. Sahud, and J. P. Quinn. 2005. First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-β-lactamase (VIM-2) in the United States. Antimicrob. Agents Chemother. 49:3538-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ploy, M.-C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rådström, P., O. Sköld, G. Swedberg, J. Flensburg, P. H. Roy, and L. Sundström. 1994. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J. Bacteriol. 176:3257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 17.Tennstedt, T., R. Szczepanowski, I. Krahn, A. Puhler, and A. Schluter. 2005. Sequence of the 68,869 bp IncP-1alpha plasmid pTB11 from a waste water treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements. Plasmid 53:218-238. [DOI] [PubMed] [Google Scholar]
- 18.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]