Abstract
Rates of metallo-β-lactamase and 16S rRNA methylase production were investigated in 51 imipenem-resistant Pseudomonas aeruginosa clinical isolates collected from hospitals in São Paulo, Brazil. Of them, 57% and 75% produced SPM-1 and RmtD, respectively. Of note, 51% produced both enzymes, suggesting that their coproduction is already common in this geographic area.
Treatment of infections due to Pseudomonas aeruginosa is becoming increasingly complicated by its tendency to acquire resistance to multiple classes of antimicrobials (12). The agents typically used to treat these infections include antipseudomonal penicillins, cephalosporins, carbapenems, fluoroquinolones, and aminoglycosides. The production of metallo-β-lactamases (MBLs) contributes substantially to panresistant phenotypes in P. aeruginosa because they confer resistance to all classes of β-lactam antimicrobials except aztreonam (10). Various MBLs including IMP, VIM, SPM, and GIM types have been reported for P. aeruginosa (12). Of note, SPM-type MBLs have been found only in Brazil thus far (13).
Methylation of 16S rRNA has emerged as a mechanism of high-level aminoglycoside resistance among gram-negative pathogens in recent years (3). Five such methylases, ArmA and RmtA through RmtD, have been reported to date. The most recently identified methylase is RmtD, which we reported previously for a panresistant P. aeruginosa strain isolated near São Paulo, Brazil (4). This enzyme confers high-level resistance to most aminoglycosides in clinical use. In this particular strain, the coproduction of RmtD and SPM-1 played a substantial role in the panresistant phenotype. The present study was conducted to determine the prevalence of coproduction of these enzymes among P. aeruginosa clinical isolates in Brazil.
Nonrepetitive imipenem-resistant P. aeruginosa isolates from a total of 49 patients hospitalized at seven hospitals in the São Paulo area in 2005 and 2006 were collected. One patient had two isolates with different pulsed-field gel electrophoresis (PFGE) patterns, and another patient had two isolates with identical PFGE patterns but markedly different susceptibility patterns. Thus, 51 imipenem-resistant P. aeruginosa isolates were included in the study. Sites of the specimens were as follows: 28 from urine, 8 from blood, 7 secretions from various sites, 3 from cerebrospinal fluid, 2 from catheter tip, 1 from bronchoalveolar lavage fluid, 1 from ascitic fluid, and 1 from nasal swab.
The MICs of imipenem, meropenem, aztreonam, amikacin, gentamicin, arbekacin, ciprofloxacin, and colistin were obtained using either the standard agar dilution method or Etest (AB Biodisk, Solna, Sweden) (2). The MICs of imipenem were 32 μg/ml or higher for all the isolates, verifying the high degree of resistance to this agent in this collection. For meropenem, MICs were 8 μg/ml or higher. When the phenotypic test using sodium mercaptoacetic acid was performed to detect MBL production (1), 29 of 51 isolates (57%) yielded a positive result.
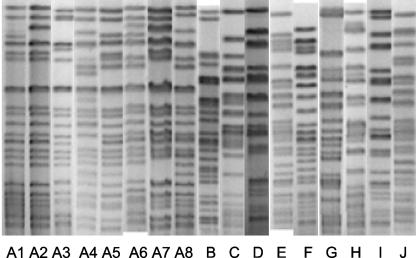
PFGE was performed with SpeI (New England Biolabs, Beverly, MA) as the restriction enzyme for the genomic DNA. Electrophoresis was performed with the CHEF III DR system (Bio-Rad, Hercules, CA). The pulses were increased linearly from 5.3 to 34.9 s for 24 h at 14°C. Seventeen pulsotypes were observed among the 51 isolates by PFGE. Eight of the pulsotypes comprising 32 isolates from six hospitals were possibly related to each other (“pulsotype A,” consisting of subtypes A1 through A8), whereas the other nine pulsotypes were unrelated (pulsotypes B through J) according to criteria described previously by Tenover et al. (Table 1 and Fig. 1) (14). Pulsotype A was thus the most prevalent genotype in hospitals in this area.
TABLE 1.
Characteristics of study isolates
| Pulsotype and subtype | Total no. of isolates | Hospital(s) | No. of isolates (%)
|
|
|---|---|---|---|---|
| blaSPM positive | rmtD positive | |||
| A | 32 | b, c, d, e, f, g | 29 (91) | 29 (91) |
| 1 | 12 | b, c, e, f, g | 11 | 12 |
| 2 | 10 | b, d, g | 9 | 8 |
| 3 | 1 | g | 1 | 1 |
| 4 | 3 | b | 2 | 3 |
| 5 | 3 | b | 3 | 3 |
| 6 | 1 | b | 1 | 1 |
| 7 | 1 | b | 1 | 1 |
| 8 | 1 | b | 1 | 0 |
| B | 1 | a | 0 (0) | 0 (0) |
| C | 5 | b | 0 (0) | 0 (0) |
| D | 1 | a | 0 (0) | 0 (0) |
| E | 7 | b | 0 (0) | 7 (100) |
| F | 1 | g | 0 (0) | 0 (0) |
| G | 1 | b | 0 (0) | 0 (0) |
| H | 1 | d | 0 (0) | 1 (100) |
| I | 1 | a | 0 (0) | 0 (0) |
| J | 1 | b | 0 (0) | 1 (100) |
FIG. 1.
PFGE patterns of the study isolates. The pulsotypes were aligned using Bionumerics software version 4.0 (Applied Maths, Sint-Martens-Latem, Belgium).
PCR was conducted to detect the MBL gene blaSPM (6) and the 16S rRNA methylase gene rmtD. PCR for blaIMP-1, blaIMP-2, and blaVIM-2 was performed for all the blaSPM-negative isolates (11). For rmtD, the following primers were used to produce a 401-bp amplicon: rmtD-F (5′-CGGCACGCGATTGGGAAGC-3′) and rmtD-R (5′-CGGAAACGATGCGACGAT-3′). The thermal cycle conditions included an initial denaturation step at 96°C for 5 min followed by 30 cycles at 96°C for 30 s, 55°C for 30 s, and 72°C for 1 min, with a final extension step at 72°C for 5 min. All the 29 isolates that produced a positive phenotypic test for MBL production yielded an amplicon consistent with blaSPM. When representative amplicons from different PFGE pulsotypes were sequenced, their deduced amino acid sequences matched that of SPM-1 within the amplicons (15). PCR results for blaIMP-1, blaIMP-2, and blaVIM-2 were all negative. These results suggested that SPM-1 is indeed the most predominant MBL among P. aeruginosa isolates in this geographic area. The median MIC of imipenem was greater than 256 μg/ml for blaSPM-positive isolates and 64 mg/ml for blaSPM-negative isolates (Table 2). For rmtD, 38 isolates (75%) yielded an amplicon. Deduced amino acid sequences of representative amplicons from different PFGE profiles were identical to that of RmtD within the amplicons. The MICs of arbekacin, amikacin, and gentamicin for the rmtD-positive isolates were always greater than 256 μg/ml. Of the 29 blaSPM-positive isolates, 26 were positive for rmtD as well. Thus, the prevalence of coproduction of SPM-1 and RmtD in this collection of imipenem-resistant P. aeruginosa isolates was 51% (26/51), and that among the SPM-1-producing isolates reached 90% (26/29). All of the blaSPM-positive isolates belonged to pulsotype A. Most of the rmtD-positive strains were also observed within pulsotype A, whereas some belonged to pulsotypes E, H, and J. These results suggest that the rmtD and blaSPM genes are spreading in São Paulo hospitals mostly by means of interhospital transmission of strains belonging to pulsotype A.
TABLE 2.
MICs of imipenem, meropenem, aztreonam, amikacin, ciprofloxacin, and colistin in the presence or absence of blaSPM
| blaSPM status (no. of isolates) | Antibiotic | No. of isolates with MICs (μg/ml) of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| >256 | 256 | 128 | 64 | 32 | 16 | 8 | 4 | 2 | ≤1 | ||
| Positive (29) | Imipenem | 21 | 6 | 2 | |||||||
| Meropenem | 17 | 10 | 1 | 1 | |||||||
| Aztreonam | 5 | 19 | 5 | ||||||||
| Amikacin | 26 | 3 | |||||||||
| Ciprofloxacin | 1 | 1 | 8 | 15 | 2 | 1 | 1 | ||||
| Colistin | 12 | 8 | 7 | 2 | |||||||
| Negative (22) | Imipenem | 17 | 5 | ||||||||
| Meropenem | 2 | 10 | 9 | 1 | |||||||
| Aztreonam | 1 | 2 | 3 | 4 | 7 | 3 | 2 | ||||
| Amikacin | 12 | 4 | 1 | 4 | 1 | ||||||
| Ciprofloxacin | 4 | 3 | 3 | 7 | 1 | 1 | 3 | ||||
| Colistin | 2 | 3 | 11 | 5 | 1 | ||||||
When the MICs of aminoglycosides were stratified according to the PCR results for rmtD, the sensitivities of arbekacin, amikacin, and gentamicin MICs greater than 256 μg/ml in predicting the presence of rmtD were all 100% (Table 3). On the other hand, the specificities of their MICs equal to or less than 256 μg/ml were 77%, 100%, and 54%, respectively. Therefore, amikacin was the best single agent to predict aminoglycoside resistance mediated by the production of RmtD. This result is in contrast with a previous finding with a collection of Acinetobacter sp. strains, where arbekacin displayed better specificity than amikacin (8). Although arbekacin generally retains better activity than amikacin in the presence of an aminoglycoside-modifying enzyme among gram-negative bacteria, this advantage may be lost when multiple modifying enzymes with different substrate specificities are produced simultaneously in a bacterium (5). Both agents are also known to be substrates of efflux pumps in P. aeruginosa (9). A combination of these resistance mechanisms may thus lead to high aminoglycoside MICs even in the absence of 16S rRNA methylase. The MICs of imipenem, meropenem, aztreonam, amikacin, ciprofloxacin, and colistin were then stratified depending on the PCR results for blaSPM (Table 2). Higher MICs of the carbapenems and lower MICs of aztreonam were observed in blaSPM-positive isolates, as expected, reflecting the substrate specificities of the MBL (10). High-level amikacin resistance was more frequent in blaSPM-positive isolates due to the frequent coproduction of RmtD. MICs of ciprofloxacin were variable regardless of the presence or absence of blaSPM. Some isolates with elevated colistin MICs (8 to 16 μg/ml) were observed in both groups, which differed from a previous study that reported 100% susceptibility to this agent for imipenem-resistant P. aeruginosa isolates in Brazil (6).
TABLE 3.
MICs of aminoglycosides in the presence or absence of rmtD
| rmtD status (no. of isolates) | Aminoglycoside | No. of isolates with MIC (μg/ml) of:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| >256 | 256 | 128 | 64 | 32 | 16 | 8 | 4 | ||
| Positive (38) | Arbekacin | 38 | |||||||
| Amikacin | 38 | ||||||||
| Gentamicin | 38 | ||||||||
| Negative (13) | Arbekacin | 3 | 2 | 1 | 6 | 1 | |||
| Amikacin | 4 | 1 | 7 | 1 | |||||
| Gentamicin | 6 | 1 | 3 | 3 | |||||
In summary, our study demonstrates that the coproduction of SPM-1 and RmtD is a common phenomenon observed in half of the imipenem-resistant P. aeruginosa isolates in hospitals in São Paulo, Brazil. A recent nationwide surveillance study from Brazil reported that as many as 37% of P. aeruginosa isolates recovered in Brazilian hospitals were resistant to imipenem (7). Furthermore, it was also reported that MIC90s of amikacin, tobramycin, and gentamicin against P. aeruginosa were all greater than 256 μg/ml. Our findings, coupled with those surveillance data, suggest that high-level aminoglycoside resistance mediated by the production of 16S rRNA methylase may already be disseminated in Brazilian hospitals.
Acknowledgments
This study was supported in part by a Fogarty International Center Global Infectious Diseases Research Training Program grant from the National Institutes of Health (grant 5D43TW006592; principal investigator, Lee H. Harrison). Y.D. was supported in part by National Institutes of Health grant T32 AI007333.
Footnotes
Published ahead of print on 18 June 2007.
REFERENCES
- 1.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and M. Goto. 2000. Convenient test for screening metallo-β-lactamase-producing gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 38:40-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Doi, Y., and Y. Arakawa. 2007. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45:88-94. [DOI] [PubMed] [Google Scholar]
- 4.Doi, Y., D. de Oliveira Garcia, J. Adams, and D. L. Paterson. 2007. Coproduction of novel 16S rRNA methylase RmtD and metallo-β-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob. Agents Chemother. 51:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi, Y., J. Wachino, K. Yamane, N. Shibata, T. Yagi, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Spread of novel aminoglycoside resistance gene aac(6′)-Iad among Acinetobacter clinical isolates in Japan. Antimicrob. Agents Chemother. 48:2075-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gales, A. C., L. C. Menezes, S. Silbert, and H. S. Sader. 2003. Dissemination in distinct Brazilian regions of an epidemic carbapenem-resistant Pseudomonas aeruginosa producing SPM metallo-β-lactamase. J. Antimicrob. Chemother. 52:699-702. [DOI] [PubMed] [Google Scholar]
- 7.Kiffer, C., A. Hsiung, C. Oplustil, J. Sampaio, E. Sakagami, P. Turner, C. Mendes, and the MYSTIC Brazil Group. 2005. Antimicrobial susceptibility of gram-negative bacteria in Brazilian hospitals: the MYSTIC Program Brazil 2003. Braz. J. Infect. Dis. 9:216-224. [DOI] [PubMed] [Google Scholar]
- 8.Lee, H., D. Yong, J. H. Yum, K. H. Roh, K. Lee, K. Yamane, Y. Arakawa, and Y. Chong. 2006. Dissemination of 16S rRNA methylase-mediated highly amikacin-resistant isolates of Klebsiella pneumoniae and Acinetobacter baumannii in Korea. Diagn. Microbiol. Infect. Dis. 56:305-312. [DOI] [PubMed] [Google Scholar]
- 9.Mao, W., M. S. Warren, A. Lee, A. Mistry, and O. Lomovskaya. 2001. MexXY-OprM efflux pump is required for antagonism of aminoglycosides by divalent cations in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:2001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 11.Pitout, J. D., D. B. Gregson, L. Poirel, J. A. McClure, P. Le, and D. L. Church. 2005. Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J. Clin. Microbiol. 43:3129-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossolini, G. M., and E. Mantengoli. 2005. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin. Microbiol. Infect. 11(Suppl. 4):17-32. [DOI] [PubMed] [Google Scholar]
- 13.Sader, H. S., A. O. Reis, S. Silbert, and A. C. Gales. 2005. IMPs, VIMs and SPMs: the diversity of metallo-β-lactamases produced by carbapenem-resistant Pseudomonas aeruginosa in a Brazilian hospital. Clin. Microbiol. Infect. 11:73-76. [DOI] [PubMed] [Google Scholar]
- 14.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]