Abstract
The rate of nonsusceptibility of penicillin-resistant Streptococcus pneumoniae strains to ceftriaxone increased significantly in Taiwan in 2005. Approximately 90% of the ceftriaxone-nonsusceptible isolates were found to be of four major serotypes (serotypes 6B, 14, 19F, and 23F). Seven amino acid alterations in the penicillin-binding protein 2B transpeptidase-encoding region specifically contributed to the resistance.
Extended-spectrum cephalosporins are important antibiotics in the treatment of invasive infections caused by penicillin-resistant Streptococcus pneumoniae, and ceftriaxone and cefotaxime are incorporated into guidelines for the empirical treatment of community-acquired pneumonia in hospitalized patients (8). Resistance to extended-spectrum cephalosporins requires sequential and multiple mutations of target genes. We report on an increase in the rate of ceftriaxone resistance among penicillin-resistant S. pneumoniae isolates in Taiwan.
The records for S. pneumoniae clinical isolates in Chang Gung Memorial Hospital and Chang Gung Children's Hospital from 2000 to 2005 were reviewed. Isolates with the same antibiogram that were obtained from the same patient within 30 days were regarded as repetitive and were removed from use for the statistical analysis.
All isolates were cultured and identified by standard methods (11). Antimicrobial susceptibility was determined by a broth microdilution method, and the results were interpreted according to the breakpoints for MICs suggested by the Clinical and Laboratory Standards Institute (CLSI) (2). The CLSI definition of ceftriaxone susceptibilities is different between isolates from cerebrospinal fluid (CSF) (susceptible, MIC ≤0.5 μg/ml; intermediately resistant, 1 μg/ml; resistant, ≥2 μg/ml) and isolates from other specimens (susceptible, ≤1 μg/ml; intermediately resistant, 2 μg/ml; resistant, ≥4 μg/ml). Isolates with intermediate or full resistance were classified as nonsusceptible.
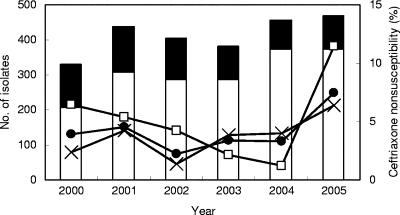
Among 3,033 pneumococcal isolates collected from 2000 to 2005, 785 were recovered from sterile sites (blood, pleural fluid, CSF, ascitic fluid, tissue, and synovial fluid), including 40 from CSF. The average annual number of isolates was 506 (maximum, 573 in 2005). The rate of penicillin nonsusceptibility increased significantly from an average of 73.5% from 2000 to 2004 to 78.5% in 2005 (P < 0.05). Before 2004, the rate of ceftriaxone nonsusceptibility was 2.2 to 4.6% (average, 3.5%), but it increased significantly to 7.4% in 2005 (P < 0.0005) (Fig. 1). The rate of ceftriaxone nonsusceptibility increased more significantly among isolates derived from sterile sites (4.2% from 2000 to 2004 to 11.5% in 2005; P < 0.005) than among those derived from nonsterile sites (3.3% from 2000 to 2004 to 6.4% in 2005; P < 0.01) (Fig. 1). The proportion of pneumococcal isolates with ceftriaxone nonsusceptibility also increased significantly among penicillin-resistant isolates (5.7% from 2000 to 2004 to 13.7% in 2005; P < 0.00001). Among strains showing intermediate resistance to penicillin, only one with intermediate resistance to ceftriaxone and the another strain that was fully resistant were identified in 2001. From 2000 to 2005, more ceftriaxone-nonsusceptible isolates were derived from pediatric patients (patients <18 years old; 5.6%) than from adults (3.5%) (P < 0.01).
FIG. 1.
Increasing rates of ceftriaxone resistance among penicillin-resistant Streptococcus pneumoniae isolates in Taiwan. The annual numbers of pneumococcal isolates from different body sites (open bars, nonsterile sites; closed bars, sterile sites) and the rapid increase in the rates of ceftriaxone nonsusceptibility among all isolates (•), isolates from nonsterile sites (×), and, notably, isolates from sterile sites (□) in 2005 are shown. Ceftriaxone nonsusceptibility was found almost exclusively among the penicillin-resistant isolates, and a rapid increase in the rate of such resistance was observed in 2005.
A total of 459 isolates that were collected from sterile sites from 2004 to 2005, when the rate of ceftriaxone resistance started to rise, were subjected to serotyping and genotyping (14, 15). Among the 441 typeable isolates, 24 different serotypes/serogroups were identified. Serotype 14 was the most common (24.6%), followed by serotypes 23F (21.1%), 6B (17.9%), 19F (14.4%), and 3 (4.8%). Four serotypes (serotypes 6B, 14, 19F, and 23F) accounted for 89.1% (326/366) of the penicillin-nonsusceptible isolates and 90% (27/30) of the ceftriaxone-nonsusceptible isolates. The remaining three ceftriaxone-resistant isolates were nontypeable. Most of the serotype 3 isolates (20/22; 90.9%) were susceptible to penicillin. The seven-valent conjugate vaccine contains capsular polysaccharides of serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F. These seven serotypes and the potentially cross-reactive serotypes/serogroups (serotypes/serogroups 6A, 6F, 9, 18, 19B, 19C, 23A, and 23B) accounted for 86.3% of the isolates examined.
Pulsed-field gel electrophoresis (PFGE) analysis revealed 110 unique genotypes among the 459 pneumococcal isolates. There were 13 genotypes among the 30 ceftriaxone-nonsusceptible isolates. The PFGE patterns of 19 (63.3%) isolates matched those of the predominant genotypes identified among the four common serotypes. Different serotypes could share the same genotype, suggesting that capsular transformation could have occurred among these isolates. Compared to the available international clones, genotypes P (serotype 14), F (serotype 19F), and B (serotype 23F) identified among our isolates were nearly identical to the established patterns of the England14, Taiwan19F, and Spain23F clones, respectively (data not shown) (7).
S. pneumoniae has five high-molecular-weight penicillin-binding proteins (PBPs), three of which (PBP 1A, PBP 2B, and PBP 2X) are involved in β-lactam resistance (10). A total of 46 isolates were analyzed by PCR and sequencing of the amino acid sequences of PBP 1A (719 amino acids), PBP 2B (685 amino acids), and PBP 2X (750 amino acids) (12). The PCR products were sequenced by using the sequencing primers shown in Table 1. According to the susceptibilities to penicillin and ceftriaxone, the 46 isolates could be placed into one of three groups: penicillin and ceftriaxone susceptible (9 isolates), penicillin nonsusceptible but ceftriaxone susceptible (12 isolates), and penicillin and ceftriaxone nonsusceptible (25 isolates). The PBP genes of strain R6 (GenBank accession nos. AE008414.1, AE008520.1, and AE008411.1) were used for comparison. EpiInfo software (version 6.04; CDC, Stone Mountain, Ga.) was used for statistical analysis. The chi-square test or Fisher's exact test was used when appropriate. A difference was considered statistically significant when the two-tailed P value was <0.05.
TABLE 1.
Primers used for sequencing
| Sequencing target | Primer sequence |
|---|---|
| pbp1a | 5′-825TGT CGG TCA TCA TAT AGG C843-3′ |
| 5′-1827TTA CCT CAG TTA GCC TTG CT1808-3′ | |
| pbp2b | 5′-1238CCA CGC TTT CCA TAT TGC CA1257-3′ |
| 5′-1966ATT ACA AGC AGT TCA GCC CGT G1945-3′ | |
| 5′-1238TGG CAA TAT GGA AAG CGT GG1219-3′ | |
| pbp2x | 5′-1673CCA GGT AGC ATC TCC CAT1656-3′ |
| 5′-915ATG CCA ATA TGA TGT CTA TC934-3′ | |
| 5′-1358GAC CGA CCT TTG ATG CAG AT1377-3′ |
Amino acid alterations were found in various sites among the three PBPs; 57 amino acid alterations in PBP 1A, 17 in PBP 2B, and 75 in PBP 2X were significantly associated with the penicillin nonsusceptibility. Furthermore, 7 amino acid alterations in PBP 2B were found to be significantly associated with the ceftriaxone nonsusceptibility: A320S, A327S, E338T, S362A, D369N, K371E, and E374D (P < 0.05 for all comparisons).
This study documents an alarming increase in the rate of ceftriaxone nonsusceptibility among S. pneumoniae isolates, especially among those causing invasive diseases. In Asia, a multinational study involving 11 countries reported an overall penicillin nonsusceptibility rate of 52.4%, which is among the highest in the world (13). However, ceftriaxone resistance was found in only 0.9% of the non-CSF isolates and 4.1% of the CSF isolates (13). Nevertheless, cases of the failure of treatment with both ceftriaxone and cefotaxime for pneumococcal meningitis were reported in the early 1990s (3, 9). The development and further dissemination of ceftriaxone resistance in S. pneumoniae observed in this study therefore have serious implications for public health, as well as for the care of individual patients.
Clinical isolates of S. pneumoniae from Asian countries, including Taiwan, are largely confined to a limited number of serotypes, most notably, 6B, 9V, 14, 19F, and 23F (13). In this study, we found a positive correlation between the pneumococcal serotypes and the proportions resistant to penicillin and ceftriaxone, suggesting that in Taiwan an existing selection pressure has been driving the development of ceftriaxone resistance among the penicillin-resistant clones. Some of the ceftriaxone-resistant strains showed genotypes similar to those of specific multidrug-resistant clones, such as Spain23F, England14, and Taiwan19F. Given the high potential for these resistant clones to evolve, reducing the selection pressure by limiting the unnecessary use of antimicrobial agents in Taiwan and other countries can never be overemphasized.
S. pneumoniae developed penicillin resistance through a complex process involving alterations of PBPs 1A, 2B, and 2X (1, 5, 6). Some reports indicated that for high-level resistance to extended-spectrum cephalosporins, only altered PBPs 1A and 2X are required (3, 9). The present study, however, demonstrated that altered PBP 2B was also involved in the development of ceftriaxone nonsusceptibility in pneumococci. It was shown that a block of divergent sequence of the PBP 2B transpeptidase-encoding region between nucleotides 1200 and 1500 containing a series of 11 amino acid substitutions was associated with penicillin resistance. Another block in PBP 2B (nucleotide 950 to 1150) that contained 7 amino acid alterations contributed significantly to ceftriaxone nonsusceptibility among penicillin-resistant isolates. A very recent study that used an in vitro PBP binding assay also showed that in penicillin- and ceftriaxone-resistant isolates, the affinity of ceftriaxone to PBP 2B was much lower than that to other PBPs (PBPs 1A, 1B, 2X, 2A, and 3), suggesting that PBP 2B may be a primary target for ceftriaxone (4). Alterations in PBP 2B thus likely play a vital role in the development of penicillin and ceftriaxone resistance. However, an alteration in PBP 2B alone would presumably not dictate the final level of penicillin or ceftriaxone resistance in pneumococci. The final level of resistance might be dependent on the collective action of multiple altered PBPs.
The increase in the rate of ceftriaxone-resistant pneumococci in Taiwan is worrisome; however, our data indicated that 90% of the isolates with ceftriaxone resistance belonged to the serotypes covered by the conjugate vaccine. We suggest that the use of the pneumococcal conjugate vaccine be implemented in developing countries as soon as possible, especially in settings with a high prevalence of resistance.
Acknowledgments
We are indebted to Ron Dagan, David Greenberg, and Nurith Porat, Pediatric Infectious Disease Unit, Soroka University Medical Center, Ben-Gurion University of the Negev, Beer-Sheva, Israel, for their kind help in serotyping some of the clinical isolates and to Jae-Hoon Song, Asian Network for Surveillance of Resistant Pathogens (ANSORP), Seoul, Korea, for providing us S. pneumoniae strains belonging to different international epidemic clones, namely, Kor 55 (Korea6B), SL P6 (Poland6B), HK P46 (England14), HK P60 (England14), Kor 51 (Taiwan19F), HK P38 (Spain23F), and HK P65 (Spain23F).
This work was supported by a grant (grant CMRPG450011 to C.-H. Chiu) from the Chang Gung Memorial Hospital.
Footnotes
Published ahead of print on 25 June 2007.
REFERENCES
- 1.Barcus, V. A., K. Ghanckar, M. Yeo, T. J. Coffey, and C. G. Dowson. 1995. Genetics of high level penicillin resistance in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol. Lett. 126:299-303. [DOI] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing, 16th informational supplement. CLSI document M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Coffey, T. J., M. Daniels, L. K. McDougal, C. G. Dowson, F. C. Tenover, and B. G. Spratt. 1995. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 39:1306-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies, T. A., W. Shang, K. Bush, and R. K. Flamm. 2006. Activity of doripenem and comparators against Streptococcus pneumoniae US clinical isolates with defined substitutions in penicillin-binding proteins (PBP) 1A, PBP 2B, and PBP 2X, abstr. C1-0038. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 5.du Plessis, M., E. Bingen, and K. P. Klugman. 2002. Analysis of penicillin-binding protein genes of clinical isolates of Streptococcus pneumoniae with reduced susceptibility to amoxicillin. Antimicrob. Agents Chemother. 46:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karlowsky, J. A., M. E. Jones, D. C. Draghi, and D. F. Sahm. 2003. Clinical isolates of Streptococcus pneumoniae with different susceptibilities to ceftriaxone and cefotaxime. Antimicrob. Agents Chemother. 47:3155-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko, K. S., and J. H. Song. 2004. Evolution of erythromycin-resistant Streptococcus pneumoniae from Asian countries that contains erm(B) and mef(A) genes. J. Infect. Dis. 190:739-747. [DOI] [PubMed] [Google Scholar]
- 8.Mandell, L. A., J. G. Bartlett, S. F. Dowell, T. M. File, Jr., D. M. Musher, and C. G. Whitney. 2003. Update of practice guidelines for the management of community-acquired pneumonia in immunocompetent adults. Clin. Infect. Dis. 37:1405-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz, R., C. G. Dowson, M. Daniels, T. J. Coffey, C. Martin, R. Hakenbeck, and B. G. Spratt. 1992. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 6:2461-2465. [DOI] [PubMed] [Google Scholar]
- 10.Nagai, K., T. A. Davies, M. R. Jacobs, and P. C. Appelbaum. 2002. Effects of amino acid alterations in penicillin-binding proteins (PBPs) 1a, 2b, and 2x on PBP affinities of penicillin, ampicillin, amoxicillin, cefditoren, cefuroxime, cefprozil, and cefaclor in 18 clinical isolates of penicillin-susceptible, -intermediate, and -resistant pneumococci. Antimicrob. Agents Chemother. 46:1273-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruoff, K. L. 1995. Streptococcus, p. 299-307. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. American Society for Microbiology, Washington, DC.
- 12.Sanbongi, Y., T. Ida, M. Ishikawa, Y. Osaki, H. Kataoka, T. Suzuki, K. Kondo, F. Ohsawa, and M. Yonezawa. 2004. Complete sequences of six penicillin-binding protein genes from 40 Streptococcus pneumoniae clinical isolates collected in Japan. Antimicrob. Agents Chemother. 48:2244-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song, J. H., S. I. Jung, K. S. Ko, N. Y. Kim, J. S. Son, H. H. Chang, H. K. Ki, W. S. Oh, J. Y. Suh, K. R. Peck, N. Y. Lee, Y. Yang, Q. Lu, A. Chongthaleong, C. H. Chiu, M. K. Lalitha, J. Perera, T. T. Yee, G. Kumarasinghe, F. Jamal, A. Kamarulzaman, N. Parasakthi, P. H. Van, C. Carlos, T. So, T. K. Ng, and A. Shibl. 2004. High prevalence of antimicrobial resistance among clinical Streptococcus pneumoniae isolates in Asia (an ANSORP study). Antimicrob. Agents Chemother. 48:2101-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorensen, U. B. 1993. Typing of pneumococci by using 12 pooled antisera. J. Clin. Microbiol. 31:2097-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]