Abstract
Staphylococcus epidermidis is a major cause of nosocomial infections because of its ability to form biofilms on the surface of medical devices. Only a few antibacterial agents are relatively active against biofilms, and rifampin, a transcription inhibitor, ranks among the most effective molecules against biofilm-related infections. Whether this efficacy is due to advantageous structural properties of rifampin or to the fact that the RNA polymerase is a favorable target remains unclear. In an attempt to answer this question, we investigated the action of different transcription inhibitors against S. epidermidis biofilm, including the newest synthetic transcription inhibitors. This comparison suggests that most of the antibiotics that target the RNA polymerase are active on S. epidermidis biofilms at concentrations close to their MICs. One of these compounds, CBR703, despite its high MIC ranks among the best antibiotics to eradicate biofilm-embedded bacteria.
Many bacteria organize in biofilms in their natural environment to protect themselves from hostile conditions. The medical community is now aware that biofilms are involved in many serious infections and that it is very difficult to eradicate them (11). Indeed, bacterial biofilms are inherently resistant to antibiotics and host defenses. Various mechanisms have been proposed to explain why very few molecules are active against biofilms: the penetration of antibiotics can be limited by the protective matrix that enclose bacteria, and/or antibiotic activity is altered by the phenotypic heterogeneity of biofilm-embedded bacteria (7). However, the presence in the biofilms of a high frequency of persister bacteria that do not grow or die in the presence of the antibiotic might be the cause of these recalcitrant infections (15).
S. epidermidis is a biofilm-forming pathogen and is responsible for a significant amount of nosocomial device-related infections (21, 29). Rifampin, in combination with other antibiotics, is frequently used to treat these infections. This molecule is able to penetrate the protective exopolysaccharidic matrix but fails to eradicate the whole biofilm, even when administered at a high concentration (31). Rifampin is also one of the most hydrophobic agents used in chemotherapy, and its efficiency might be due to its physicochemical properties (e.g., hydrophobicity or size), which allow it to penetrate the biofilm matrix and perform its bactericidal activity. It is also possible that its intracellular target, the RNA polymerase (RNAP), is of particular importance for biofilm survival.
To further address this question, we have investigated the behavior of different antibacterial inhibitors of RNAP from seven structurally unrelated families on S. epidermidis biofilms: rifampin and two recently commercialized and more hydrophobic analogs, rifapentine and rifaximin; streptolydigin (25); lipiarmycin (5, 28), which is currently in a clinical trial under the name of OPT-80 (1); two putative transcription inhibitors (22, 23) from the pyrrothines family, thiolutin and holomycin; the recently described synthetic molecules CBR703 and its more active analogue, CBR64 (4, 18); SB2 (2, 9); and a ureidothiophene (2) (Fig. 1). The activity on biofilms was determined by an ATP-counting bioluminescence assay (8) used in several studies of antibacterial activity against S. epidermidis biofilms (18, 19).
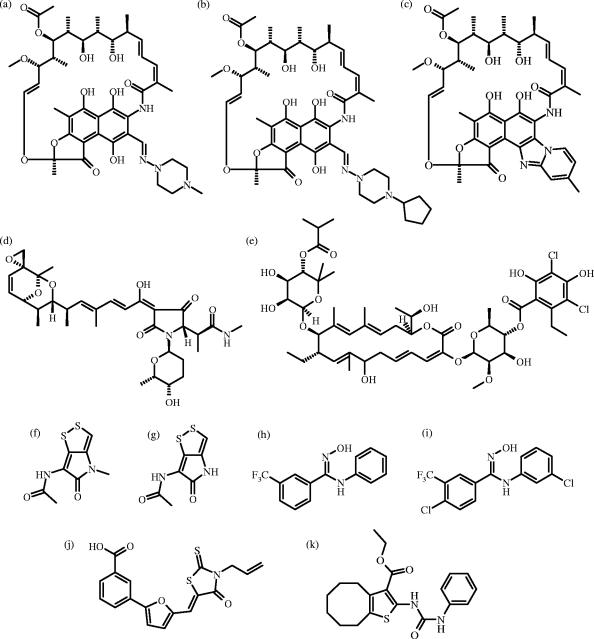
FIG. 1.
Chemical structures of rifampin (a), rifapentine (b), rifaximin (c), streptolydigin (d), lipiarmycin (e), thiolutin (f), holomycin (g), CBR703 (h), CBR64 (i), SB2 (j), and ureidothiophene (k).
MATERIALS AND METHODS
Bacterial strain.
S. epidermidis RP62A (CIP 105777) was used for its ability to colonize solid supports such as plastic culture dishes and catheters.
Antibiotics and experimental compounds.
Rifampin was purchased from Sigma-Aldrich. Rifapentine and rifaximin were purchased from Sequoia Research Products, Pangbourne, United Kingdom. Streptolydigin, holomycin, and thiolutin were purchased from Sourcon-Padena, Tübingen, Germany. Lipiarmycin was produced according to the method of Talpaert et al. (28). CBR703 was purchased from Maybridge, Tintagel, United Kingdom. CBR64 was synthesized according to the method of Li et al. (17; L. Li et al., 19 July 2001, World Intellectual Property Organization). SB2 was synthesized according to the method of Leonetti et al. (J.-P. Leonetti et al., 10 March 2005, World Intellectual Property Organization) (14). The ureidothiophene was purchased from Chembridge, San Diego, CA.
Planktonic MIC and MBC determination procedures.
The MIC and minimal bactericidal concentration (MBC) were determined as recommended by the Clinical and Laboratory Standards Institute (20). Antibiotics were tested at final concentrations (prepared from serial twofold dilutions) ranging from 0.1 × 10−4 to 2 × 10−4 μg/ml for rifampin, rifapentine, and rifaximin and from 200 to 0.4 μg/ml for streptolydigin, lipiarmycin, thiolutin, holomycin, CBR703, CBR64, SB2, and ureidothiophene. The MIC was defined as the lowest antibiotic concentration that yielded no visible growth. The test medium was Mueller-Hinton broth (MHB), and the inoculum was 5 × 105 CFU/ml. The inoculated microplates were incubated at 37°C for 24 h before being read. The MBCs were established by extending the MIC procedure to the evaluation of bactericidal activity. After 24 h, 10-μl portions were drawn from the wells, serially diluted, and then spotted onto MHB-agar plates. The plates were incubated at 37°C. The MBC was read 24 h later as the lowest concentration of antibiotic that resulted in 0.1% survival in the subculture. All of the experiments were done in triplicate.
Transcription inhibition assay.
The inhibitory effects of the RNAP inhibitors on transcription were measured according to the method of Gualtieri et al. (10) on toluene-permeabilized S. epidermidis cells. The template for these reactions was the endogenous S. epidermidis DNA, and the concentration necessary to inhibit 50% of the [5-3H]UTP incorporation was calculated by using the LSW data analysis tool (MDL, San Leandro, CA). All of the experiments were done in triplicate.
Growth of biofilms in 96-well polystyrene microtiter plates.
The wells of a black 96-well polystyrene microtiter plate (Greiner Bio-One, Frickenhausen, Germany) were filled with 0.1-ml aliquots of S. epidermidis inoculum (107 CFU/ml) in MHB, and the plate was incubated for 24 h at 37°C. Each well was rinsed three times with 0.2 ml of sterile water to remove nonadherent bacteria. Subsequently, 0.1 ml of MHB containing the desired antibiotic concentration was added to each well, and the plate was incubated at 37°C for 24 h without shaking. After the challenge, the plate was washed three times with 0.2 ml of sterile water to remove nonadherent bacteria, and 10 μl of dimethyl sulfoxide was added to each well to lyse the bacteria. The plate was shaken for 5 min, and 90 μl of ATP-counting bioluminescence buffer (25 mM HEPES, 24 mM MgCl2, 2 mM EDTA, 2 mM dithiothreitol, 60 μg of bovine serum albumin/ml, 20 μg of luciferase/ml, 0.6 mM d-luciferin [pH 7.2]5) was added to each well before reading with a luminometer (Chameleon; Hidex, Turku, Finland) (8). All of the experiments were done in triplicate.
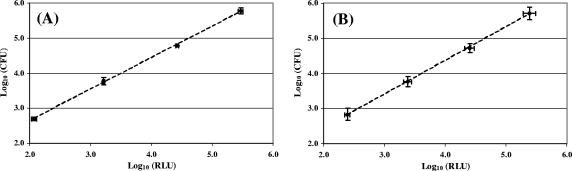
Correlation between luminescence and the amount of biofilm-embedded bacteria.
Previous studies have shown that the relationship between bacterial ATP measured by luminometry and the corresponding bacterial count was linear and that dimethyl sulfoxide was a good ATP extractant (8, 18). To establish a calibration curve for our experiments, 24-h biofilms and 48-h biofilms of S. epidermidis were grown in wells of a microtiter plate as described above. Culture medium was replaced by fresh MHB after 24 h of growth in the case of 48 h-biofilms. After incubation at 37°C for 24 h or 48 h, the biofilms were rinsed three times with 0.2 ml of sterile water to remove nonadherent bacteria. The biofilms were resuspended in 0.1 ml of sterile water by sonication using a Branson 450 sonifier with a microtip (four times for 2 s each time, 10% of the maximal amplitude). Each bacterial suspension was serially diluted in sterile water, and 10 μl of this serial dilution were drawn from the wells to be measured by bioluminescence while the number of bacteria in the wells was estimated by classical plate counting. Luminescence was measured in arbitrary relative luminescence units (RLU). All of the experiments were done in triplicate. The same procedure was applied with a 48-h biofilm (Fig. 2). Measures by bioluminescence and by plate counting allowed us to establish the following correlation equations:
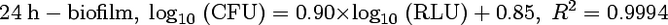 |
(1) |
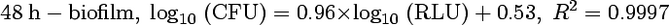 |
(2) |
Since no significant difference was observed between these equations, equation 1 was used to assess the amount of embedded bacteria within challenged and unchallenged biofilms.
FIG. 2.
Correlation curves between the ATP-counting luminescence values and the amount of biofilm-embedded S. epidermidis bacteria. (A) 24-h biofilm; (B) 48-h biofilm. The mean value ± the standard error is shown.
Quantification of persister bacteria within the biofilm.
Ciprofloxacin was used by Keren et al. to assess the amount of persister bacteria within planktonic cultures of Staphylococcus aureus (13). Thus, we have used ciprofloxacin to quantify persister cells in S. epidermidis biofilms. A S. epidermidis 24-h biofilm grown under the conditions described above was challenged with ciprofloxacin at 20 μg/ml in MHB for 24 h. After the challenge the bacterial count was estimated by the ATP-bioluminescence method used for the transcription inhibitors. This experiment was done in triplicate.
RESULTS
As expected, rifampin, rifapentine, and rifaximin—three commercial molecules—were the most active in terms of transcription inhibition. The ureidothiophene inhibited RNAP at a concentration of <1 μg/ml and the other less-evolved molecules—lipiarmycin, streptolydigin, CBR, and SB2—inhibited the RNAP at reasonable concentrations (1 to 5 μg/ml) but were far less active in the transcription assay than ansamycins. The pyrrothines thiolutin and holomycin did not inhibit RNAP. When tested on planktonic S. epidermidis, all of these compounds except ureidothiophene were bactericidal at concentrations ranging from 1× to 4× MIC (Table 1).
TABLE 1.
Biological activity of the studied molecules on bacterial growth and transcription
| Inhibitora | MIC (μg/ml)b | MBC (μg/ml) | MBC/MIC | Mean IC50(μg/ ml) ± SE |
|---|---|---|---|---|
| RIF | 0.006 | 0.006 | 1 | 0.04 ± 0.02 |
| RFP | 0.012 | 0.025 | 2 | 0.08 ± 0.03 |
| RFX | 0.012 | 0.025 | 2 | 0.07 ± 0.03 |
| STL | 50.0 | 100 | 2 | 2.30 ± 0.80 |
| LIP | 50.0 | 50.0 | 1 | 1.40 ± 0.80 |
| THL | 6.25 | 12.5 | 2 | >100 |
| HOL | 12.5 | 50.0 | 4 | >100 |
| CBR703 | 100 | 100 | 1 | 4.20 ± 1.00 |
| CBR64 | 25.0 | 25.0 | 1 | 2.30 ± 0.70 |
| SB2 | 12.5 | 25.0 | 2 | 2.50 ± 0.80 |
| URT | 6.25 | >100 | >18 | 0.50 ± 0.30 |
RIF, rifampin; RFP, rifapentine; RFX, rifaximin; STL, streptolydigin; LIP, lipiarmycin; THL, thiolutin; HOL, holomycin; URT, ureidothiophene.
MIC values were determined by the broth microdilution method.
cInhibition of transcription directed by S. epidermidis RNAP.
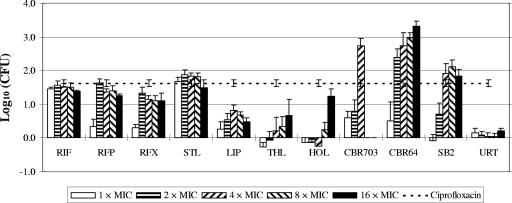
These molecules were evaluated on biofilms with a concentration range from 1× to 16× MIC, except for CBR703, which was poorly soluble at more than 4× MIC. The mean bacterial count was 6.55 ± 0.02 log10 CFU/well. The different transcription inhibitors challenged against S. epidermidis were evaluated for their ability to decrease the bacterial count within the biofilm and were compared to rifampin (Fig. 3).
FIG. 3.
Difference of S. epidermidis counts in log10 (CFU/well) between unchallenged and challenged 24-h biofilms as measured by bioluminescence. Challenged biofilms were exposed for 24 h to the transcription inhibitors at concentrations ranging from 1× MIC to 16× MIC. The mean bacterial amount within the unchallenged biofilm was 6.55 ± 0.02 log10 CFU/well. The dashed line represents the difference of counts after challenge with ciprofloxacin at 20 μg/ml for 24 h. The mean value of the bacterial count ± the standard error is given. RIF, rifampin; RFP, rifapentine; RFX, rifaximin; STL, streptolydigin; LIP, lipiarmycin; THL, thiolutin; HOL, holomycin; URT, ureidothiophene.
As expected from a bacteriostatic agent, ureidothiophene had no effect on biofilms at concentrations up to 16× MIC. All of the other bactericidal inhibitors, with the exception of CBR and the pyrrothines thiolutin and holomycin, seemed to reach a limit of activity at a concentration of 4× MIC.
Due to its very low MIC, rifampin was more active in the present study than all of the other molecules in terms of concentration, but rifampin failed to decrease the bacterial count by more than 2 log10. The other ansamycines, rifapentine and rifaximin, presented activities that were comparable to rifampin against S. epidermidis biofilms. These molecules were able to decrease the bacterial count by about 1.5 log10 at concentrations equal to their respective MBCs. These more hydrophobic analogs did not present any significant advantage over rifampin regarding biofilm eradication.
However, at concentrations close to their MICs, less-evolved molecules are as efficient or even more efficient than rifampin. When normalized on the basis of the MIC, the most active compounds against S. epidermidis biofilms appeared to be the CBR-type compounds. CBR703 and mainly CBR64 succeeded at decreasing the bacterial amount by about 3 log10 at a concentration as low as 4× MIC. These values are very high in terms of concentration. Nevertheless, this series of synthetic compounds can be optimized to decrease the MICs, as exemplified by the lower value of the MIC of CBR64 compared to the parent compound CBR703 (Table 1). CBR compounds presented a clear advantage over the reference inhibitor rifampin. They were the only structural family to display such a property in the present study since the others did not kill biofilm-embedded S. epidermidis far beyond the threshold of 2 log10.
The second most interesting molecule was the other synthetic molecule, SB2. It was able to decrease the bacterial count on a biofilm by more than 1.8 log10 at a concentration of 4× MIC.
Streptolydigin appeared to be efficient on biofilms at a concentration close to its MIC, and this activity was comparable to the activity of the ansamycine family in terms of reduction of the bacterial count. Lipiarmycin was only able to decrease the bacterial count by 0.5 to 1 log10 at a concentration of 2× MIC. The pyrrothines, thiolutin, and holomycin were not or weakly active against biofilms at concentration below 16× MIC.
Ciprofloxacin was used by Keren et al. (13) to quantify the level of persister cells within Staphylococcus aureus planktonic cultures. We extended this procedure to S. epidermidis biofilms and, after 24 h of exposure to the antibiotic, surviving bacteria—purported to be persister cells—accounted for 4.93 ± 0.10 log10, i.e., ca. 2% of the biofilm-embedded bacteria (Fig. 3).
DISCUSSION
The molecules we have tested can be classified into three categories. (i) Rifampin, rifapentine, rifaximin (27), CBR (4), lipiarmycin (10), and streptolydigin (30) are molecules that inhibit transcription in vitro and whose target has been genetically validated by the isolation of resistant mutants in the genes coding for the RNA polymerase. (ii) SB2 (2, 9) and ureidothiophene (3) are molecules that inhibit transcription in vitro, but no genetic data has proven that the target is the RNA polymerase. (iii) Holomycin and thiolutin have recurrently been suspected to affect transcription elongation in growing bacteria, but no effects have been observed in vitro (22, 23).
All of the inhibitors investigated here demonstrated bactericidal action on S. epidermidis with the exception of the ureidothiophene. However, we have observed (P. Villain-Guillot, unpublished results) that ureidothiophene properties strongly depend on the concentration of the inoculum: ureidothiophene was bactericidal with an inoculum of 5 × 104 CFU/ml. The bacteriostatic property of ureidothiophene was unexpected from an RNAP inhibitor since transcription inhibitors are expected to be bactericidal, possibly by triggering programmed cell death in bacteria (6, 24, 26).
The comparison of lipiarmycin, rifampin and its more hydrophobic analogs rifapentine and rifaximin, and the recent synthetic molecules (SB and CBR) allowed us to conclude that the physicochemical properties (e.g., hydrophobicity or size) are not the only significant factors that can explain the advantage of rifampin over other antibacterial agents on biofilms. The molecules rifampin, rifapentine, rifaximin, streptolydigin, lipiarmycin, CBR, and SB2 share common properties: they are bactericidal on planktonic S. epidermidis, and they are active against biofilms at concentrations close to their respective MICs. Lipiarmicyn's moderate activity might be due to its pH dependence (5), and pH is not homogeneous within a biofilm (12).
Interestingly, thiolutin and holomycin do not share similar properties with the RNAP inhibitors whose target has been genetically validated. In spite of their bactericidal properties, thiolutin and holomycin were active on biofilms at concentrations higher than 8× MIC in contrast to the other transcription inhibitors. Thiolutin and holomycin may fail to penetrate the protective matrix of the biofilm, but it should be noted that the mechanism of action of these molecules remains unclear. For instance, both molecules preferentially inhibit transcription in a whole-cell assay, whereas they do not inhibit RNAP in vitro (22), suggesting an indirect mechanism of action and that RNAP may not be their primary target.
Despite the good activity of rifampin, rifapentine, rifaximin, streptolydigin, and SB2, these molecules could not eradicate the whole biofilm at concentrations up to 16× MIC. This is probably due to the presence of “backup” persister bacteria within the biofilm. These persister bacteria may be a leading cause of the recalcitrance of biofilm-related pathologies (16). Persister bacteria accounted for 2% of the bacterial population in our study, and it seems that rifampin, rifapentine, rifaximin, streptolydigin, and SB2 could not exceed a threshold of 2 log10 in bacterial count reduction. These results are intriguing, but they do not permit us to conclude that only persister bacteria remain in the biofilm after the challenge with these antibacterial agents. However, CBR-type molecules could eradicate biofilm-embedded bacteria clearly above 2 log10. Thus, CBR703 and CBR64 have a bactericidal activity against persister bacteria.
The bacterial RNAP is a good target for the development of new bactericidal antibiotics, as is highlighted by the recent publication of three new synthetic inhibitors (2-4). In the present study we have compared clinically used molecules with excellent pharmacological properties to natural molecules and the latest synthetic inhibitors of transcription to determine whether this enzyme is truly of interest to generate new drugs that are active on biofilms. It appears that the relative hydrophobicity of some of these compounds or their high MICs will preclude their immediate clinical use, but this comparison suggests that all of the antibiotics that directly target the RNA polymerase, such as rifampin, rifapentine, rifaximin, streptolydigin, lipiarmycin, and CBRs, are active on S. epidermidis biofilms at concentrations close to their MICs. These data support the use of RNAP as a target for generating new drugs that are active against biofilms. The CBR-type compounds are very promising in this regard. It is difficult to predict whether these molecules will successfully evolve toward antibacterial drugs, but their ability to affect biofilm-embedded persister bacteria may be very advantageous for curing biofilm-related infections.
Acknowledgments
We are grateful to J. Martinez and M. Amblard for hosting us in their lab during the chemical synthesis of CBR64 and SB2.
P.V.-G. was supported by a postgraduate fellowship from DGA. This study was supported by institutional funds from the Centre National de la Recherche Scientifique and the Grant Biosécurité 2004.
Footnotes
Published ahead of print on 2 July 2007.
REFERENCES
- 1.Ackermann, G., B. Loffler, D. Adler, and A. C. Rodloff. 2004. In vitro activity of OPT-80 against Clostridium difficile. Antimicrob. Agents Chemother. 48:2280-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.André, E., L. Bastide, S. Michaux-Charachon, A. Gouby, P. Villain-Guillot, J. Latouche, A. Bouchet, M. Gualtieri, and J. P. Leonetti. 2006. Novel synthetic molecules targeting the bacterial RNA polymerase assembly. J. Antimicrob. Chemother. 57:245-251. [DOI] [PubMed] [Google Scholar]
- 3.Arhin, F., O. Belanger, S. Ciblat, M. Dehbi, D. Delorme, E. Dietrich, D. Dixit, Y. Lafontaine, D. Lehoux, J. Liu, G. A. McKay, G. Moeck, R. Reddy, Y. Rose, R. Srikumar, K. S. Tanaka, D. M. Williams, P. Gros, J. Pelletier, T. R. Parr, Jr., and A. R. Far. 2006. A new class of small molecule RNA polymerase inhibitors with activity against rifampicin-resistant Staphylococcus aureus. Bioorg. Med. Chem. 14:5812-5832. [DOI] [PubMed] [Google Scholar]
- 4.Artsimovitch, I., C. Chu, A. S. Lynch, and R. Landick. 2003. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science 302:650-654. [DOI] [PubMed] [Google Scholar]
- 5.Coronelli, C., R. J. White, G. C. Lancini, and F. Parenti. 1975. Lipiarmycin, a new antibiotic from Actinoplanes. II. Isolation, chemical, biological and biochemical characterization. J. Antibiot. 28:253-259. [DOI] [PubMed] [Google Scholar]
- 6.Engelberg-Kulka, H., R. Hazan, and S. Amitai. 2005. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 118:4327-4332. [DOI] [PubMed] [Google Scholar]
- 7.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34-40. [DOI] [PubMed] [Google Scholar]
- 8.Gracia, E., A. Fernandez, P. Conchello, J. L. Alabart, M. Perez, and B. Amorena. 1999. In vitro development of Staphylococcus aureus biofilms using slime-producing variants and ATP-bioluminescence for automated bacterial quantification. Luminescence 14:23-31. [DOI] [PubMed] [Google Scholar]
- 9.Gualtieri, M., L. Bastide, P. Villain-Guillot, S. Michaux-Charachon, J. Latouche, and J. P. Leonetti. 2006. In vitro activity of a new antibacterial rhodanine derivative against Staphylococcus epidermidis biofilms. J. Antimicrob. Chemother. 58:778-783. [DOI] [PubMed] [Google Scholar]
- 10.Gualtieri, M., P. Villain-Guillot, J. Latouche, J. P. Leonetti, and L. Bastide. 2006. Mutation in the Bacillus subtilis RNA polymerase β′ subunit confers resistance to lipiarmycin. Antimicrob. Agents Chemother. 50:401-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 12.Hunter, R. C., and T. J. Beveridge. 2005. Application of a pH-sensitive fluoroprobe (C-SNARF-4) for pH microenvironment analysis in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:2501-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keren, I., N. Kaldalu, A. Spoering, Y. Wang, and K. Lewis. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13-18. [DOI] [PubMed] [Google Scholar]
- 14.Leonetti, J.-P., E. André, and L. Bastide. 2006. Antibiotic thiazolidines. Patent WO 2005/020990.
- 15.Lewis, K. 2005. Persister cells and the riddle of biofilm survival. Biochemistry 70:267-274. [DOI] [PubMed] [Google Scholar]
- 16.Lewis, K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48-56. [DOI] [PubMed] [Google Scholar]
- 17.Li, L., X. Chen, P. Fan, J. T. Mihalic, and S. Cutler. 2001. Patent WO 2001/51456.
- 18.Monzon, M., C. Oteiza, J. Leiva, and B. Amorena. 2001. Synergy of different antibiotic combinations in biofilms of Staphylococcus epidermidis. J. Antimicrob. Chemother. 48:793-801. [DOI] [PubMed] [Google Scholar]
- 19.Monzon, M., C. Oteiza, J. Leiva, M. Lamata, and B. Amorena. 2002. Biofilm testing of Staphylococcus epidermidis clinical isolates: low performance of vancomycin in relation to other antibiotics. Diagn. Microbiol. Infect. Dis. 44:319-324. [DOI] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M07-A6. NCCLS, Wayne, PA.
- 21.O'Gara, J. P., and H. Humphreys. 2001. Staphylococcus epidermidis biofilms: importance and implications. J. Med. Microbiol. 50:582-587. [DOI] [PubMed] [Google Scholar]
- 22.Oliva, B., A. O'Neill, J. M. Wilson, P. J. O'Hanlon, and I. Chopra. 2001. Antimicrobial properties and mode of action of the pyrrothine holomycin. Antimicrob. Agents Chemother. 45:532-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Neill, A., B. Oliva, C. Storey, A. Hoyle, C. Fishwick, and I. Chopra. 2000. RNA polymerase inhibitors with activity against rifampin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 44:3163-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinehart, K. L., Jr., J. R. Beck, D. B. Borders, W. W. Epstein, T. H. Kinstle, L. D. Spicer, D. Krauss, and A. C. Button. 1963. Structure of streptolydigin. Antimicrob. Agents Chemother. 161:346-348. [PubMed] [Google Scholar]
- 26.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Severinov, K., M. Soushko, A. Goldfarb, and V. Nikiforov. 1993. Rifampicin region revisited. New rifampicin-resistant and streptolydigin-resistant mutants in the beta subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 268:14820-14825. [PubMed] [Google Scholar]
- 28.Talpaert, M., F. Campagnari, and L. Clerici. 1975. Lipiarmycin: an antibiotic inhibiting nucleic acid polymerases. Biochem. Biophys. Res. Commun. 63:328-334. [DOI] [PubMed] [Google Scholar]
- 29.von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2:677-685. [DOI] [PubMed] [Google Scholar]
- 30.Yang, X., and C. W. Price. 1995. Streptolydigin resistance can be conferred by alterations to either the β or β′ subunits of Bacillus subtilis RNA polymerase. J. Biol. Chem. 270:23930-23933. [DOI] [PubMed] [Google Scholar]
- 31.Zheng, Z., and P. S. Stewart. 2002. Penetration of rifampin through Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 46:900-903. [DOI] [PMC free article] [PubMed] [Google Scholar]