Abstract
We used an assay to measure quinolone sensitivity as a shift in the position of the cleavage-religation equilibrium. This assay was found to be useful in identifying the primary target of a quinolone drug and assessing the effect of quinolone resistance-conferring mutations.
Topoisomerases are responsible for altering the linking number of DNA (1, 11, 15). Type II topoisomerases, including DNA gyrase and topoisomerase IV (Topo IV), change the linking number of DNA in steps of two by introducing a double-strand break into duplex DNA, passing another segment of the duplex through the break, and then resealing the broken strands (1, 15). Under normal circumstances, the covalent topoisomerase-DNA complex is a fleeting catalytic intermediate. The steady-state level of the covalent topoisomerase-DNA complex depends on the cleavage-religation equilibrium, which is equivalent to the ratio of the cleavage and religation rates. If the equilibrium is shifted to either stimulate cleavage or inhibit religation by a topoisomerase inhibitor, the covalent topoisomerase-DNA complex can persist on the DNA as if the topoisomerase were trapped in a topoisomerase-drug-DNA ternary complex. This class of topoisomerase inhibitors is often referred to as “topoisomerase poisons” because of their unique mode of action (5, 10, 12, 14). Topoisomerase poisons include quinolone antibacterial drugs and anticancer drugs, such as etoposide and camptothecin. Quinolone drugs target and poison both DNA gyrase and Topo IV (2, 14).
Quinolone resistance-conferring mutations arise rapidly in gyrase and Topo IV genes (2, 14). Some mutations affect not only the quinolone sensitivity but also the biochemical properties of a topoisomerase, which makes it difficult to determine the effect of a mutation. For instance, the E84K mutation in ParC affects the catalytic activity of Topo IV (6) and slows down the growth rate of Escherichia coli (9). As a result, much greater amounts of the mutant Topo IV, relative to the wild-type enzyme, were required to perform commonly used in vitro assays. The slow growth would also influence in vivo measurements of quinolone sensitivity. Thus, the effect of a quinolone resistance-conferring mutation on the functional activity of a topoisomerase could make the determination of its quinolone sensitivity problematic.
Since the mechanism of quinolone drugs is to shift the position of the cleavage-religation equilibrium (2, 5, 12), it seemed reasonable to determine quinolone sensitivity in a manner more consistent with the action of quinolone drugs. We decided to modify a DNA cleavage assay and measure the quinolone sensitivity of a topoisomerase by comparing the steady-state level of the cleavage-religation equilibrium in the presence of drug with that in the absence of drug. Specific activities for spontaneous and quinolone-induced DNA cleavage by topoisomerases were measured, and quinolone sensitivity was defined as the ratio of the cleavage activity in the presence of an excess concentration of norfloxacin to that in the absence of norfloxacin. We used this assay to compare the quinolone sensitivities of E. coli and Staphylococcus aureus gyrases and Topo IVs and assess the effects of quinolone resistance-conferring mutations.
E. coli and S. aureus gyrases and Topo IVs, as well as the quinolone-resistant mutant enzymes ParC S80L Topo IV and ParC E84K Topo IV, were described previously (6, 7). Negatively supercoiled plasmid pBR322 DNA and norfloxacin were purchased from New England BioLabs and Sigma, respectively.
A DNA cleavage assay was performed according to a protocol similar to that described by Fortune and Osheroff (4). Briefly, reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 8 at 23°C), 10 mM MgCl2, 10 mM dithiothreitol, 50 μg/ml bovine serum albumin, 1 mM ATP, 5 μg/ml tRNA, 300 ng of pBR322 DNA, and the indicated amounts (as a tetramer) of various topoisomerases were incubated at 37°C for 10 min in the absence or presence of 50 μM norfloxacin (this is an excess amount to induce the maximum level of DNA cleavage by all of the topoisomerases used in this study [data not shown]). Sodium dodecyl sulfate was added to a concentration of 1%, and the reaction mixtures were further incubated at 37°C for 5 min. EDTA and proteinase K were then added to 25 mM and 100 μg/ml, respectively, and the incubation was continued for an additional 15 min at 37°C. The DNA products were purified by extraction with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol) and then analyzed by electrophoresis through vertical 1.2% agarose gels (14 by 10 by 0.3 cm) at 2 V/cm for 12 h in TAE buffer (50 mM Tris-HCl [pH 7.9 at 23°C], 40 mM sodium acetate, 1 mM EDTA) that contained 0.5 μg/ml ethidium bromide. After destaining in water, gels were photographed and quantified with an Eagle Eye II system (Stratagene).
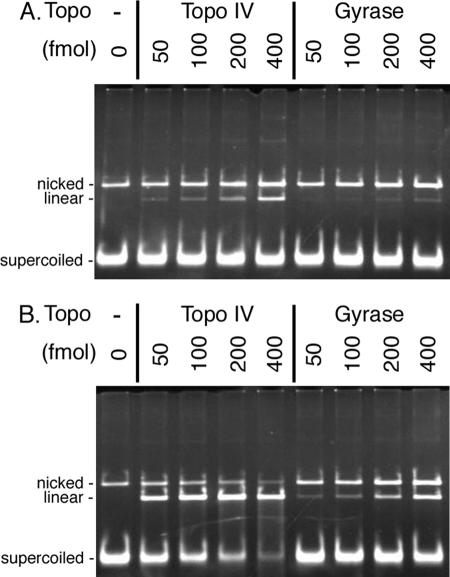
Each topoisomerase was titrated in the absence or presence of norfloxacin to measure its spontaneous and quinolone-induced cleavage activity. An example of the results of this assay is shown in Fig. 1. DNA cleavage was monitored by the conversion of negatively supercoiled DNA to linear DNA. Specific activities (amounts of linear DNA generated per unit amount of a topoisomerase) for spontaneous and quinolone-induced DNA cleavage were determined by using the data where the increase in the linear DNA generated by a topoisomerase as a function of topoisomerase concentration showed a linear response. The total amount of the full-length DNA molecules (negatively supercoiled, nicked, and linear DNA molecules) was also monitored to be sure that it remained constant (reduction was less than 10%). Quinolone sensitivity was defined as follows: quinolone sensitivity = (specific activity of a topoisomerase for quinolone-induced DNA cleavage)/(specific activity of a topoisomerase for spontaneous DNA cleavage).
FIG. 1.
Determination of the quinolone sensitivities of S. aureus gyrase and Topo IV. The DNA cleavage activity of either S. aureus gyrase or S. aureus Topo IV was measured in the absence (A) or presence (B) of 50 μM norfloxacin with the negatively supercoiled plasmid DNA (100 fmol as a molecule) as the substrate. Amounts of topoisomerases as tetramers are indicated.
Although DNA gyrase and Topo IV are both cellular targets of the quinolone antibacterial drugs (2, 14), a quinolone drug has its primary target in vivo. Interestingly, DNA gyrase in gram-negative bacteria and Topo IV in gram-positive bacteria are often found to be the primary targets. For instance, DNA gyrase is the primary target of quinolone drugs in E. coli (9, 10) whereas Topo IV becomes the primary target in S. aureus (3). A study has shown, however, that each quinolone drug appears to have a preferred target and the target selection can be altered by changes in quinolone structure (13). Thus, it is not clear what determines the primary target in cells. Quinolone sensitivities of gyrase and Topo IV can be readily measured by standard in vitro assays. The relative biochemical sensitivity (or the primary biochemical target), however, does not always match the identity of genes associated with the first-step mutations (or the primary target in vivo). This might be due to the distinct but complementary roles of Topo IV and gyrase in DNA replication and/or the different mechanisms used to repair gyrase and Topo IV lesions (8, 9).
Quinolone sensitivities of gyrase and Topo IV are likely to be among the key factors that determine the primary target. Our assay allowed us to measure the quinolone sensitivities of E. coli and S. aureus gyrases and Topo IVs under the same conditions, despite their different biochemical properties. We measured the spontaneous and quinolone-induced cleavage activities of these topoisomerases and determined their quinolone sensitivities (Table 1). E. coli gyrase was much more sensitive to norfloxacin than E. coli Topo IV was, whereas S. aureus gyrase was less sensitive to norfloxacin than S. aureus Topo IV was. A dramatic difference in quinolone sensitivity between E. coli gyrase and E. coli Topo IV (18-fold, in contrast to the only 2.5-fold difference between the S. aureus topoisomerases) might explain why Topo IV is such a poor target of quinolone drugs in E. coli (2, 9). These results demonstrated that, in fact, primary targets of norfloxacin in either E. coli or S. aureus were more sensitive to norfloxacin than secondary targets were. Thus, our assay for quinolone sensitivity appeared to be a useful method to identify the primary target of a quinolone drug in the cell.
TABLE 1.
Quinolone sensitivities of E. coli and S. aureus type II topoisomerases
| Topoisomerase | Cleavage activitya
|
Quinolone sensitivity | |
|---|---|---|---|
| Without norfloxacin | With norfloxacin | ||
| E. coli DNA gyrase | 0.0057 | 0.39 | 68.4 |
| E. coli Topo IV | 0.14 | 0.53 | 3.79 |
| S. aureus DNA gyrase | 0.0049 | 0.049 | 10.0 |
| S. aureus Topo IV | 0.017 | 0.42 | 24.7 |
Nanograms of linear DNA generated per femtomole (as a tetramer) of a topoisomerase.
Next, we measured the drug sensitivities of two quinolone-resistant mutant Topo IVs, ParC S80L Topo IV and ParC E84K Topo IV, which exhibit distinct biochemical properties (6). As shown in Table 2, either the S80L or the E84K mutation in E. coli ParC reduced the quinolone sensitivity of E. coli Topo IV. These results coincided well with previous observations that both the S80L and E84K mutations in ParC confer quinolone resistance on Topo IV (2, 6, 14). Thus, the assay described here appeared to be useful in assessing the effect of quinolone resistance-conferring mutations on a topoisomerase, irrespective of their effect on the catalytic activity of the topoisomerase.
TABLE 2.
Effect of quinolone resistance-conferring mutations on E. coli Topo IV
| Topoisomerase | Cleavage activitya
|
Quinolone sensitivity | |
|---|---|---|---|
| Without norfloxacin | With norfloxacin | ||
| Wild-type Topo IV | 0.14 | 0.53 | 3.79 |
| ParC S80L Topo IV | 0.25 | 0.49 | 1.96 |
| ParC E84K Topo IV | 0.14 | 0.17 | 1.21 |
Nanograms of linear DNA generated per femtomole (as a tetramer) of a topoisomerase.
We used a pair of in vitro DNA cleavage assays to measure the spontaneous and quinolone-induced DNA cleavage activities of a topoisomerase and determine its quinolone sensitivity as the ratio of the specific activity of the topoisomerase for quinolone-induced DNA cleavage to that for spontaneous DNA cleavage. This assay was developed to measure the quinolone sensitivity based on the mechanism of quinolone drugs and thus more accurately reflect their cytotoxicity. This assay appears to be a useful method to determine the primary target of a quinolone drug in various bacteria, as well as to assess the effects of mutations on the quinolone sensitivity of topoisomerases.
Acknowledgments
We thank Lisa Oppegard for critical reading of the manuscript.
Funding to pay the publication charges for this article was provided by the University of Minnesota Medical School.
Footnotes
Published ahead of print on 2 July 2007.
REFERENCES
- 1.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 2.Drlica, K., and M. Malik. 2003. Fluoroquinolones: action and resistance. Curr. Top. Med. Chem. 3:249-282. [DOI] [PubMed] [Google Scholar]
- 3.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 4.Fortune, J. M., and N. Osheroff. 1998. Merbarone inhibits the catalytic activity of human topoisomerase II by blocking DNA cleavage. J. Biol. Chem. 273:17643-17650. [DOI] [PubMed] [Google Scholar]
- 5.Froelich-Ammon, S. J., and N. Osheroff. 1995. Topoisomerase poisons: harnessing the dark side of enzyme mechanism. J. Biol. Chem. 270:21429-21432. [DOI] [PubMed] [Google Scholar]
- 6.Hiasa, H. 2002. The Glu-84 of the ParC subunit plays critical roles in both topoisomerase IV-quinolone and topoisomerase IV-DNA interactions. Biochemistry 41:11779-11785. [DOI] [PubMed] [Google Scholar]
- 7.Hiasa, H., M. E. Shea, C. M. Richardson, and M. N. Gwynn. 2003. Staphylococcus aureus gyrase-quinolone-DNA ternary complexes fail to arrest replication fork progression in vitro: effects of salt on the DNA binding mode and the catalytic activity of Staphylococcus aureus gyrase. J. Biol. Chem. 278:8861-8868. [DOI] [PubMed] [Google Scholar]
- 8.Khodursky, A. B., and N. R. Cozzarelli. 1998. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J. Biol. Chem. 273:27668-27677. [DOI] [PubMed] [Google Scholar]
- 9.Khodursky, A. B., E. L. Zechiedrich, and N. R. Cozzarelli. 1995. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl. Acad. Sci. USA 92:11801-11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreuzer, K. N., and N. R. Cozzarelli. 1979. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J. Bacteriol. 140:424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine, C., H. Hiasa, and K. J. Marians. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 1400:29-43. [DOI] [PubMed] [Google Scholar]
- 12.Li, T.-K., and L. F. Liu. 2001. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 41:53-77. [DOI] [PubMed] [Google Scholar]
- 13.Pan, X.-S., and L. M. Fisher. 1997. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 41:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reece, R. J., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26:335-375. [DOI] [PubMed] [Google Scholar]
- 15.Wang, J. C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. 3:430-440. [DOI] [PubMed] [Google Scholar]