Abstract
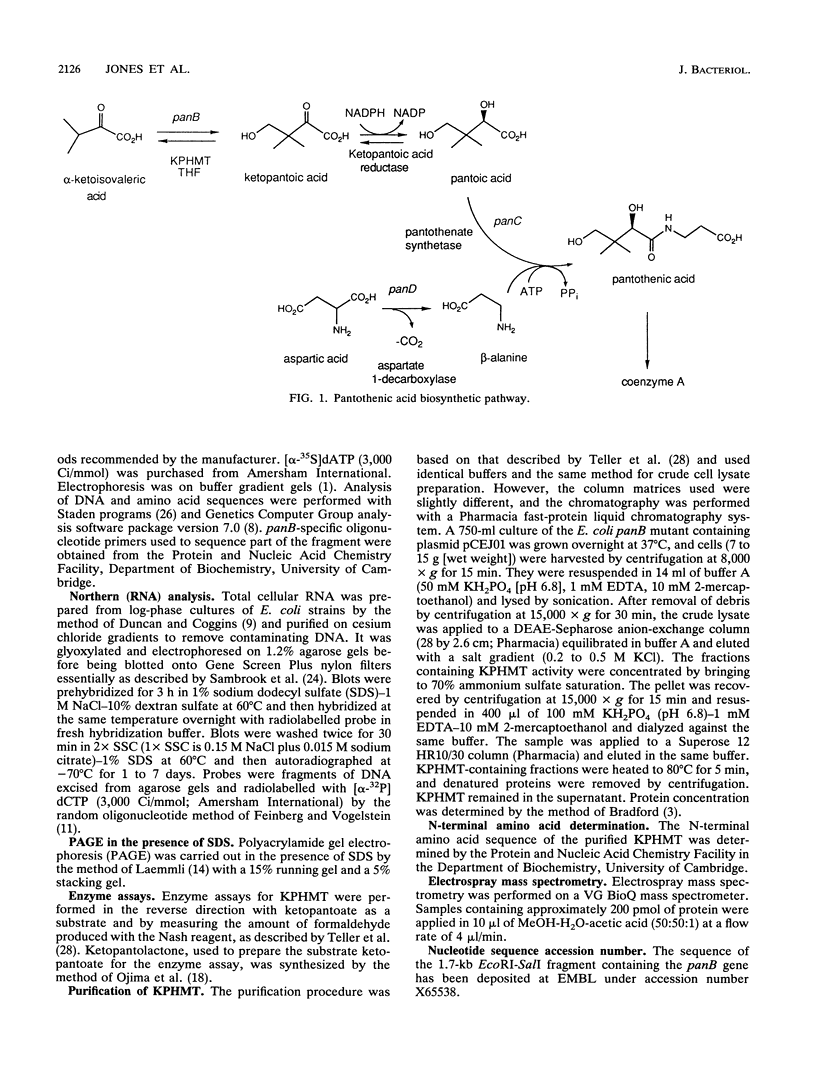
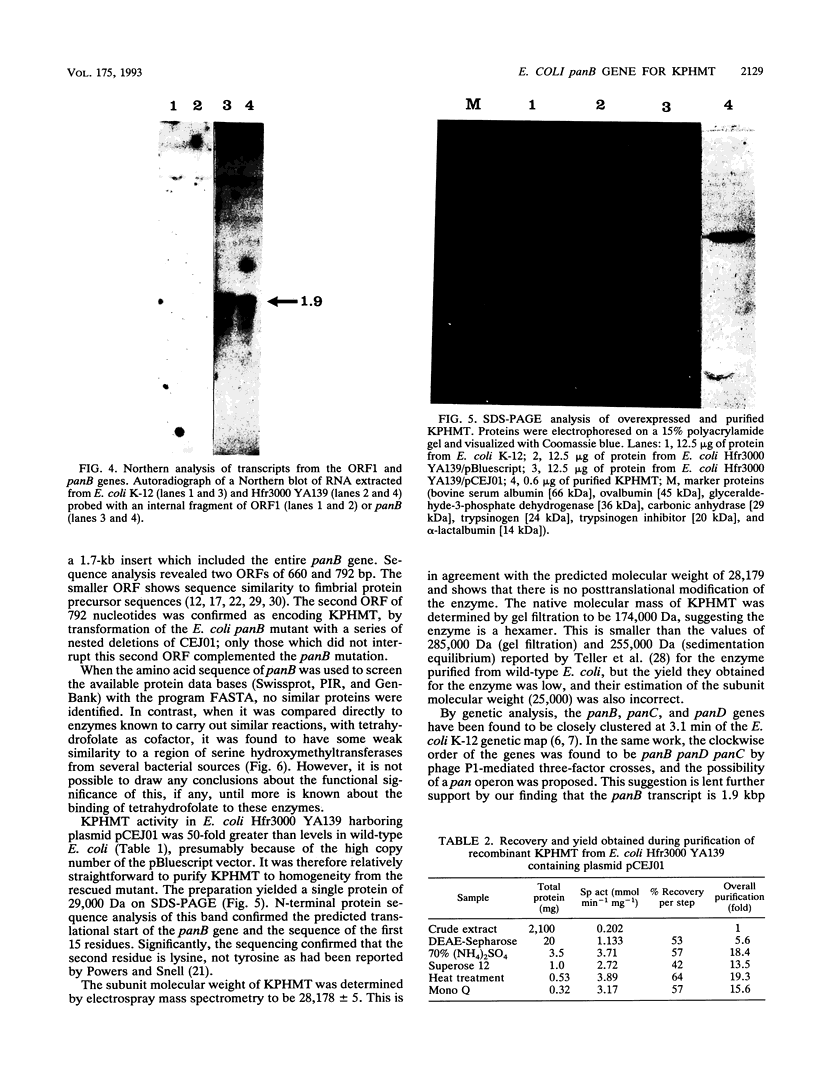
The panB gene from Escherichia coli, encoding the first enzyme of the pantothenate biosynthesis pathway, ketopantoate hydroxymethyltransferase (KPHMT), has been isolated by functional complementation of a panB mutant strain with an E. coli genomic library. The gene is 792 bp long, encoding a protein of 264 amino acids with a predicted M(r) of 28,179. The identity of the gene product as ketopantoate hydroxymethyltransferase was confirmed by purification of the enzyme protein, which was overexpressed approximately 50-fold in the mutant harboring the gene on a high-copy-number plasmid. The N-terminal amino acid sequence of the purified protein was found to be identical to that predicted from the gene sequence, as was its mass, determined by electrospray mass spectrometry. Upstream of the panB gene is an incomplete open reading frame encoding a protein of 220 amino acids, which shares sequence similarity to fimbrial precursor proteins from other bacteria. Northern (RNA) analysis showed that the panB gene is likely to be cotranscribed with at least one other gene but that this is not the putative fimbrial protein, since no transcripts for this gene could be detected.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brown G. M., Williamson J. M. Biosynthesis of riboflavin, folic acid, thiamine, and pantothenic acid. Adv Enzymol Relat Areas Mol Biol. 1982;53:345–381. doi: 10.1002/9780470122983.ch9. [DOI] [PubMed] [Google Scholar]
- Chan V. L., Bingham H. L. Complete sequence of the Campylobacter jejuni glyA gene encoding serine hydroxymethyltransferase. Gene. 1991 May 15;101(1):51–58. doi: 10.1016/0378-1119(91)90223-x. [DOI] [PubMed] [Google Scholar]
- Cronan J. E., Jr Beta-alanine synthesis in Escherichia coli. J Bacteriol. 1980 Mar;141(3):1291–1297. doi: 10.1128/jb.141.3.1291-1297.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan J. E., Jr, Littel K. J., Jackowski S. Genetic and biochemical analyses of pantothenate biosynthesis in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1982 Mar;149(3):916–922. doi: 10.1128/jb.149.3.916-922.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan K., Coggins J. R. The serC-aro A operon of Escherichia coli. A mixed function operon encoding enzymes from two different amino acid biosynthetic pathways. Biochem J. 1986 Feb 15;234(1):49–57. doi: 10.1042/bj2340049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds C. G., Smith R. D. Electrospray ionization mass spectrometry. Methods Enzymol. 1990;193:412–431. doi: 10.1016/0076-6879(90)93430-s. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gerlach G. F., Clegg S., Allen B. L. Identification and characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae. J Bacteriol. 1989 Mar;171(3):1262–1270. doi: 10.1128/jb.171.3.1262-1270.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MAAS W. K., DAVIS B. D. Pantothenate studies. I. Interference by D-serine and L-aspartic acid with pantothenate synthesis in Escherichia coli. J Bacteriol. 1950 Dec;60(6):733–745. doi: 10.1128/jb.60.6.733-745.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAAS W. K., VOGEL H. J. alpha-Ketoisovaleric acid, a precursor of pantothenic acid in Escherichia coli. J Bacteriol. 1953 Apr;65(4):388–393. doi: 10.1128/jb.65.4.388-393.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunoe Y., Nakabeppu Y., Sekiguchi M., Kawabata S., Moriya T., Amako K. Cloning and sequence of the gene encoding the major structural component of mannose-resistant fimbriae of Serratia marcescens. J Bacteriol. 1988 Aug;170(8):3567–3574. doi: 10.1128/jb.170.8.3567-3574.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M. D., Stauffer L. T., Urbanowski M. L., Stauffer G. V. Complete nucleotide sequence of the E. coli glyA gene. Nucleic Acids Res. 1983 Apr 11;11(7):2065–2075. doi: 10.1093/nar/11.7.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S. G., Snell E. E. Ketopantoate hydroxymethyltransferase. II. Physical, catalytic, and regulatory properties. J Biol Chem. 1976 Jun 25;251(12):3786–3793. [PubMed] [Google Scholar]
- Purcell B. K., Pruckler J., Clegg S. Nucleotide sequences of the genes encoding type 1 fimbrial subunits of Klebsiella pneumoniae and Salmonella typhimurium. J Bacteriol. 1987 Dec;169(12):5831–5834. doi: 10.1128/jb.169.12.5831-5834.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach S., Hennecke H. Identification of glyA as a symbiotically essential gene in Bradyrhizobium japonicum. Mol Microbiol. 1991 Jan;5(1):39–47. doi: 10.1111/j.1365-2958.1991.tb01824.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiert J. G., Urbanowski M. L., Stauffer L. T., Plamann M. D., Stauffer G. V. Nucleotide sequence of the Salmonella typhimurium glyA gene. DNA Seq. 1990;1(2):107–113. doi: 10.3109/10425179009016038. [DOI] [PubMed] [Google Scholar]
- Teller J. H., Powers S. G., Snell E. E. Ketopantoate hydroxymethyltransferase. I. Purification and role in pantothenate biosynthesis. J Biol Chem. 1976 Jun 25;251(12):3780–3785. [PubMed] [Google Scholar]
- van Die I., Bergmans H. Nucleotide sequence of the gene encoding the F72 fimbrial subunit of a uropathogenic Escherichia coli strain. Gene. 1984 Dec;32(1-2):83–90. doi: 10.1016/0378-1119(84)90035-0. [DOI] [PubMed] [Google Scholar]
- van Ham S. M., Mooi F. R., Sindhunata M. G., Maris W. R., van Alphen L. Cloning and expression in Escherichia coli of Haemophilus influenzae fimbrial genes establishes adherence to oropharyngeal epithelial cells. EMBO J. 1989 Nov;8(11):3535–3540. doi: 10.1002/j.1460-2075.1989.tb08519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]