A recent report described the sequence of a conjugative IncHI2 plasmid, pAPEC-O1-R, from an avian pathogenic extraintestinal Escherichia coli isolate (2). pAPEC-O1-R shares a large fraction of its genome with the IncHI2 plasmid R478 (1), with regions responsible for conjugative transfer, replication, and incompatibility and for conferring resistance to tellurite, silver, and copper being almost identical (2). pAPEC-O1-R also confers resistance to several antibiotics, including gentamicin, streptomycin, sulfonamides, and tetracycline, and to the antiseptic benzalkonium chloride. The genes, reported as aac(3)-VI, aadA, sul1, tetAR, and qacEΔ1, are clustered in a segment that is unique to pAPEC-O1-R and located between the tellurite and the silver resistance regions.
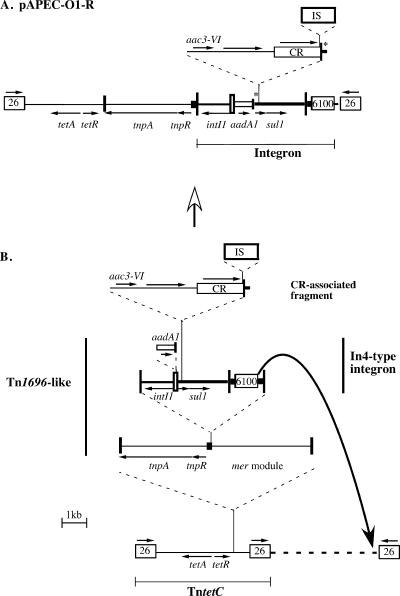
A more detailed analysis, reported here, reveals that the unique segment in pAPEC-O1-R (bp 85758 to 108613 in GenBank accession no. DQ517526) is a mosaic made up of parts of several known entities, as well as some new ones. The segment between the two IS26 (bp 87750 to 108097), which includes all of the antibiotic resistance genes, is also mosaic (Fig. 1A). The integron, an In4-type class 1 integron (6) carrying only the aadA1 gene cassette, is much smaller (bp 95259 to 107269) than indicated previously and does not include the tetracycline resistance determinant. It extends from the imperfect inverted repeat IRi (the outer boundary of the 5′-conserved segment) to IS6100, as the short segment of the tni transposition module and the imperfect inverted repeat IRt end, normally found to the right of IS6100 in the In4 family (see Fig. 1B), is missing. There is a large insertion within the 3′-conserved segment (3′-CS) of the integron that includes a new CR (rolling circle) element related to CR1 and CR3, which are also found within class 1 integrons (5). This insertion includes a short stretch from the beginning of the 3′-CS of class 1 integrons, which is duplicated in pAPEC-O1-R (Fig. 1A) and appears to have been incorporated by homologous recombination, as described previously for CR1-containing and CR3-containing class 1 integrons (3, 5). The aac(3)-VIa gene is associated with the incoming CR element, and the sequence of part of this region is almost identical to one reported previously for the aac(3)-VIa gene and an associated CR1-like region (7). The CR element is interrupted by an insertion sequence (IS) which separates the orf513-related gene from the readily recognizable ori end (5).
FIG. 1.
Structure of the multiple antibiotic resistance region on pAPEC-O1-R (A) and the proposed progenitor (B). The region shown in panel A is bp 87750 to 108091 in GenBank accession no. DQ517526. The true extent of the In4-type integron (bp 95259 to 107269) is marked below. The asterisks mark the short stretch of the 3′-conserved segment that is duplicated. Lines of various widths represent the different recognizable segments that make up these regions. The dashed horizontal line represents unknown sequence. The genes are indicated by horizontal arrows with names, if relevant. Tall vertical bars represent the inverted repeats at the boundaries of transposons and the class 1 integron. IS are represented by open boxes with the IS number inside them, and for IS26, an arrow shows the orientation. The unnumbered IS (bp 102627 to 103968 in GenBank accession no. DQ517526) lies within the ori end of the CR element and is flanked by an 8-bp duplication; its closest relative is ISCsp1 in GenBank accession no. AY639948. The new CR element is shown as a box with a vertical bar at one end representing the conserved sequence at the ori end of the CR (see reference 5), which is found at bp 103995 to 104021. The closest relative of the open reading frame in the CR is ORF2 from CR3 (87% identical). The open vertical bar represents the attI site of the integron, and the open box with short vertical bar represents the gene cassette.
The integron is in precisely the same position as In4 in Tn1696 (4) (see Fig. 1B). The tet(C) tetracycline resistance determinant lies to the left of the Tn1696 transposition module in a structure that comprises one end of the tet(C)-containing transposon found in Tn1404* (8, 9), which is flanked by two copies of IS26. The Tn1696-like entity appears to have transposed into the central region of this transposon (Fig. 1B). As the 9 bp found between IS6100 and the right-hand copy of IS26 in pAPEC-O1-R are not those expected for the integron tni fragment, it appears that the deletion shown in the figure, which removed all of the intervening segments, was initiated by IS6100 rather than IS26.
Footnotes
Published ahead of print on 25 June 2007.
REFERENCES
- 1.Gilmour, M. W., N. R. Thomson, M. Sanders, J. Parkhill, and D. E. Taylor. 2004. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52:182-202. [DOI] [PubMed] [Google Scholar]
- 2.Johnson, T. J., Y. M. Wannemeuhler, J. A. Scaccianoce, S. J. Johnson, and L. K. Nolan. 2006. Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob. Agents Chemother. 50:3929-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levings, R. S., D. Lightfoot, S. R. Partridge, R. M. Hall, and S. P. Djordjevic. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rather, P. N., P. A. Mann, R. Mierzwa, R. S. Hare, G. H. Miller, and K. J. Shaw. 1993. Analysis of the aac(3)-VIa gene encoding a novel 3-N-acetyltransferase. Antimicrob. Agents Chemother. 37:2074-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnabel, E. L., and A. L. Jones. 1999. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl. Environ. Microbiol. 65:4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokes, H. W., L. D. H. Elbourne, and R. M. Hall. 2007. Tn1403, a multiple antibiotic resistance transposon made up of three distinct transposons. Antimicrob. Agents Chemother. 51:1827-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]