Abstract
Several genotypic resistance algorithms have been proposed for quantitation of the degree of phenotypic resistance to the human immunodeficiency virus (HIV) protease inhibitor (PI) lopinavir (LPV), including the original LPV mutation score. In this study, we retrospectively evaluated 21 codons in HIV protease known to be associated with PI resistance in a large antiretroviral agent-experienced observational patient cohort, “Autorisation Temporaire d'Utilization” (ATU), to assess whether a more optimal algorithm could be derived by using virologic response data from patients treated with LPV in combination with ritonavir (LPV/r). Five of the 11 mutations constituting the LPV mutation score were not associated with a virologic response, while 4 additional mutations not included in this score demonstrated an association. Therefore, the LPV ATU score, which includes mutations at codons 10, 20, 24, 33, 36, 47, 48, 54, 82, and 84, was constructed and shown in two different types of multivariable analyses of the ATU cohort to be a better predictor of the virologic response than the LPV mutation score. The LPV ATU score was also more strongly associated with a virologic response when it was applied to independent clinical trial populations of PI-experienced patients receiving LPV/r. This study provides the basis for a new genotypic resistance algorithm that is useful for predicting the antiviral activities of LPV/r-based regimens in PI-experienced patients. The refined algorithm may be useful in making clinical treatment decisions and in refining genetic and pharmacologic methods for assessing the activity of LPV/r.
The use of potent antiretroviral therapy for the treatment of human immunodeficiency virus (HIV) type 1 (HIV-1) infection has led to a reduction in HIV-related morbidity and mortality over the past decade (6). However, the emergence of viable HIV-1 strains that are resistant to the available antiretroviral drugs has presented significant challenges to the successful long-term treatment of HIV-1 infection (7). Drug-resistant HIV-1 strains emerge when drug levels in the blood are too low to prevent viral replication but are high enough to exert selective pressure (11). Because resistance to one drug can engender cross-resistance to other drugs in the same class, the potency of subsequent antiretroviral therapy may be compromised (7, 9). Therefore, quantitative assessment of drug resistance is essential for constructing optimal drug combinations when the presence of drug-resistant HIV-1 is suspected.
Genotypic and/or phenotypic resistance testing has been widely used in clinical practice to select regimens for the treatment of HIV-1 infection. Although the results of phenotypic tests are conceptually easier to interpret, genotypic assays are generally less costly and more readily available and may provide more precise information regarding susceptibility to current antiretroviral drugs and the potential for further resistance development (genetic barrier). For these reasons, the development of genotypic algorithms has been a major focus, resulting in several HIV-1 drug resistance scores (1, 7, 17, 22, 24). However, due to the dynamic nature of the virus and the introduction of new drugs that select for different viral mutants, it is necessary to review and refine the available scores on an ongoing basis. One means of accomplishing this is to continually test the existing algorithms as new data on viral isolates become available from controlled clinical studies, observational cohort studies, and larger patient databases.
The HIV-1 protease inhibitor (PI) lopinavir (LPV) is coformulated with low-dose ritonavir (with the combination abbreviated LPV/r), resulting in enhanced plasma LPV levels due to decreased LPV metabolism through ritonavir inhibition of intestinal and hepatic cytochrome P450 3A. LPV/r has been shown to have substantial antiviral activity in patients infected with HIV-1 strains resistant to other PIs (2, 12, 23). Several genotypic resistance algorithms have been constructed to quantitate the degree of phenotypic resistance to LPV (13, 21, 25). The first algorithm was defined by the analysis of the relationship between genotypic data and phenotypic susceptibility from a limited set (n = 112) of viral isolates (13). This initial analysis led to the identification of 11 codons in the protease gene, mutations at which were associated with reduced in vitro susceptibility to LPV (LPV mutation score): wild-type amino acid L at position 10 mutated to F, I, R, or V (abbreviated L10F/I/R/V), K20M/R, L24I, M46I/L, F53L, I54L/T/V, L63P, A71I/L/T/V, V82A/F/T, I84V, and L90M, with each individual mutation assigned an equal weight. The number of these mutations present at the baseline correlated with the virologic response in multiple-PI-experienced, nonnucleoside reverse transcriptase inhibitor (NNRTI)-naive clinical trial subjects (12). Subsequently, two separate analyses of the genotypic and phenotypic relationships for subject samples (n > 1,300) from much larger databases resulted in two refined lists of mutations that correlated with in vitro resistance to LPV: Parkin et al. (21) included L10F/I/R/V, G16E, K20M/R, L24I, V32I, L33F, E34Q, K43T, M46I/L, I47A/V, G48M/V, I50V, I54A/L/M/S/T/V, K55E, L63P/T, A71I/L/T/V, G73T, T74S, V82A/F/S/T, I84V, L89I/M, and L90M, with each individual mutation assigned a weight of 1, with the exception of the I47A, I50V, I54A/M/S/T/V, and V82A/F/S mutations, which were each assigned a weight of 3; and Wang and Larder (25) included L10I, Q18Y, L24I, V32I, L33F/M, K43T, K45T, M46I/L, G48V, F53L, I54A/S/V, K55R, Q58E, A71V, I72V, G73S/T, T74S, V82A, I84V, I85V, L90M, and C95F/L, with each individual mutation assigned an equal weight.
Since these analyses were based on larger data sets, the newer algorithms included mutations that occur at relatively low frequencies. Both of these algorithms were shown to be superior to the LPV mutation score in predicting reduced in vitro susceptibility to LPV (21, 25).
The three LPV resistance algorithms discussed above were developed by examining the relationship between the genotype and in vitro phenotypic susceptibility. In theory, algorithms defined by analysis of the virologic response to a specific therapy with respect to the baseline genotype should be more clinically relevant. In order to test this possibility and to determine whether refinement of the LPV mutation score would yield a more optimal algorithm, we assessed the effects of individual mutations within the LPV mutation score on the virologic response after treatment with LPV/r and evaluated whether additional codons were associated with an impaired response to LPV/r. This assessment was performed by retrospective analysis of the association between the virologic response and the baseline HIV-1 genotype in patients receiving LPV/r in a large observational cohort.
(The data presented in this report have been presented previously [8, 14, 20].)
MATERIALS AND METHODS
Patients.
The “Autorisation Temporaire d'Utilization” (ATU; an LPV/r preregistrational usage program in France) observational cohort (n = 792) included antiretroviral agent-experienced HIV-1-infected patients for whom the baseline HIV-1 drug resistance genotype and follow-up data were available. The entry criteria initially restricted enrollment to subjects with CD4+ T-cell counts of <200 cells/mm3; the restrictions were gradually reduced to allow the use of LPV/r in a broader PI-experienced population. The data collected included the plasma HIV-1 RNA level, CD4+ T-cell count, and serious adverse events.
Virologic analyses.
Quantification of plasma HIV-1 RNA was performed by using the COBAS AMPLICOR Monitor (version 1.5) assay (Roche Diagnostics, Basel, Switzerland), which has a lower limit of detection of 400 copies/ml.
Plasma HIV-1 RNA was used for sequence analysis of the protease gene (codons 1 to 99). Plasma HIV-1 RNA was amplified by a one-step reverse transcription-PCR by using a TITAN one-tube reverse-transcription PCR kit (Boehringer Mannheim, Mannheim, Germany), followed by a nested PCR with AmpliTaq Gold (Applied Biosystems, Foster City, CA). The primers used were described previously (10, 15, 19). Direct sequencing of the PCR product was performed by using the dRhodamine Terminator cycle sequencing ready reaction kit (PE Applied Biosystems). The sequencing reaction products were analyzed on an ABI 377 genetic analyzer (PE Applied Biosystems). The sequences were analyzed by the use of Sequence Navigator software (PE Applied Biosystems) by comparing the sense and antisense strands of each fragment with the wild-type virus HXB2 sequence at codons 10, 20, 24, 30, 32, 33, 36, 46, 47, 48, 50, 53, 54, 63, 71, 73, 77, 82, 84, 88, and 90 within the HIV-1 protease domain.
Baseline characteristics.
Descriptive statistics were calculated for demographics, baseline characteristics, and treatment history. The baseline prevalence of the mutations at each codon was summarized by number and percentage.
Virologic response.
Two virologic response end points were assessed: (i) a 1.0-log10 decrease from the baseline in the plasma HIV-1 RNA level at any time, and (ii) the achievement of a plasma HIV-1 RNA level of <400 copies/ml at any time. Subjects achieving a response as defined for the second end point were considered responders for the first end point, even if they did not demonstrate a 1.0-log10 copy/ml decrease due to low baseline HIV-1 RNA levels and the lower limit of quantitation of the assay.
For each end point, the proportion of responders was summarized overall and, for each codon, for subjects with and without a mutation. Odds ratios and 95% confidence intervals (CIs) were reported. For each end point, the response was analyzed as a function of the LPV mutation score by logistic regression.
Relationship between HIV-1 genotype and virologic response.
The LPV mutation score was defined as the sum of the mutations (see the Introduction) in the protease domain derived from the genotypic-phenotypic correlations described previously (13). To assess the relationship between the HIV-1 genotype and the virologic response directly, each codon was evaluated as follows: first, for each subject, the LPV mutation score was computed while ignoring the codon of interest. Second, logistic regression analyses were used to assess the significance of a mutation at the codon of interest after adjustment for the LPV mutation score. One analysis evaluated the probability of a 1.0-log10 decrease in the plasma HIV-1 RNA level, while a second one evaluated the probability of achieving a plasma HIV-1 RNA level of <400 copies/ml. Codons demonstrating statistically significant (P < 0.05) reductions in the probability of response for both end points and codons that were nonsignificant but that exhibited evidence of a reduced likelihood of a response (odds ratio, <0.85 for both end points; i.e., a 15% reduction in the odds of a response) were retained. The number of such mutations for each subject was defined as the LPV ATU score. The relationship between the proportion of responders and the LPV ATU score for each end point was analyzed by logistic regression.
Multivariable analyses.
Two analyses were conducted to assess the significance of the two mutation scores in the context of other baseline variables. In particular, a multiple logistic regression model invoking a stepwise selection procedure (P < 0.05 to enter and stay in the model) and binary recursive partitioning (classification tree analysis [3]) were used to assess the probability of a 1.0-log10 decrease from the baseline in the plasma HIV-1 RNA level. Potential predictor variables included the LPV mutation score; the LPV ATU score; individual protease mutations; the baseline plasma HIV-1 RNA level; and measures of previous and concurrent antiretroviral medication use, including the number of prior medications by class, the number of active antiretroviral medications in the background regimen, active NNRTI use (no baseline NNRTI mutations and NNRTI use in the regimen), or new NNRTI use (no prior NNRTI use and NNRTI use in the regimen). For partitions based on continuous or ordinal variables, the algorithm used for binary recursive partitioning selects the optimal division among the values of the variable; therefore, breakpoints are not prespecified. As a sensitivity analysis, the relationship between the mutation scores and the maximum decrease from the baseline in the plasma HIV-1 RNA level was assessed by using an analysis of covariance with factors for the number of PIs used previously, the use of a new NNRTI, and the baseline plasma HIV-1 RNA level.
Application to other populations.
The LPV ATU score was applied separately to three clinical study populations of antiretroviral agent-experienced HIV-1-infected subjects who participated in one of the following previously described LPV/r clinical studies: Study 765 and Study 888 each enrolled single PI-experienced, NNRTI-naive patients to receive LPV/r with nevirapine and two nucleoside reverse transcriptase inhibitors (NRTIs) (2, 23); and Study 957 enrolled multiple-PI-experienced, NNRTI-naive patients to receive LPV/r with efavirenz and two NRTIs (12). For these analyses, a virologic response was defined as a plasma HIV-1 RNA level of <400 copies/ml at week 48 by using an intent-to-treat, dropouts-as-censored analysis. Based on the findings of Parkin and colleagues (21) that mutations at codons 47, 50, 54, and 82 may have a greater effect on LPV resistance, an exploratory weighted version of the LPV ATU score by the use of weights of 3 at codons 54 and 82 and weights of 1 at each of the other codons was calculated and applied to each of the three clinical study populations. An additional weight was not assigned at codons 47 and 50 due to the low prevalence of mutations at those codons.
Statistical software.
Analyses were conducted by using SAS (version 8.2; SAS Institute, Cary, NC), except for binary recursive partitioning, which was conducted by using CART (Salford Systems, Inc., San Diego, CA).
RESULTS
Baseline characteristics.
The ATU cohort comprised 792 subjects with a mean age of 42 years, of whom 82% were male and 53% had previously experienced an AIDS-defining illness (Centers for Disease Control and Prevention category class C [4]). The mean ± standard deviation baseline plasma HIV-1 RNA level and CD4+ T-cell count were 4.8 ± 0.76 log10 copies/ml and 178 ± 159 cells/mm3, respectively. The mean number of prior PIs used was 3.1, and 78% of the subjects were NNRTI experienced. The median (25th to 75th percentile) duration of follow-up was 14 (9 to 23) weeks.
Mutations at amino acid positions 10, 63, 71, and 90 in the protease gene were the most prevalent, occurring in more than 50% of the subjects (Table 1).
TABLE 1.
Prevalence of protease mutations at baseline and association with virologic response to LPV/r
| Codon (protease) | No. (%) with individual mutations at baseline | Subjects with 1.0-log10-copies/ml HIV-1 RNA decrease from baseline
|
Subjects achieving HIV-1 RNA levels of <400 copies/ml
|
||||
|---|---|---|---|---|---|---|---|
| Proportion responding (% with mutation vs % without mutation) | Unadjusted odds ratio (95% CI) | Adjusted odds ratioa (95% CI) | Proportion responding (% with mutation vs % without mutation) | Unadjusted odds ratio (95% CI) | Adjusted odds ratioa (95% CI) | ||
| 10 | 506 (64) | 65 vs 83 | 0.41 (0.29, 0.58) | 0.58 (0.39, 0.88) | 39 vs 62 | 0.40 (0.29, 0.53) | 0.48 (0.34, 0.68) |
| 20 | 138 (17) | 59 vs 74 | 0.50 (0.34, 0.73) | 0.63 (0.42, 0.93) | 32 vs 50 | 0.47 (0.32, 0.69) | 0.56 (0.38, 0.84) |
| 24 | 34 (4) | 56 vs 73 | 0.48 (0.24, 0.96) | 0.60 (0.30, 1.22) | 26 vs 48 | 0.39 (0.18, 0.85) | 0.48 (0.22, 1.06) |
| 30 | 27 (3) | 93 vs 71 | 5.08 (1.19, 21.6) | 3.09 (0.71, 13.4) | 74 vs 46 | 3.35 (1.40, 8.02) | 2.31 (0.95, 5.61) |
| 32 | 23 (3) | 74 vs 72 | 1.11 (0.43, 2.86) | 1.39 (0.53, 3.62) | 43 vs 47 | 0.86 (0.37, 2.00) | 1.06 (0.45, 2.47) |
| 33 | 43 (5) | 63 vs 72 | 0.64 (0.34, 1.22) | 0.79 (0.41, 1.51) | 26 vs 48 | 0.37 (0.18, 0.74) | 0.44 (0.22, 0.89) |
| 36 | 313 (40) | 66 vs 76 | 0.63 (0.46, 0.86) | 0.72 (0.52, 1.00) | 40 vs 52 | 0.61 (0.46, 0.82) | 0.68 (0.50, 0.91) |
| 46 | 342 (43) | 67 vs 76 | 0.64 (0.47, 0.87) | 0.78 (0.56, 1.07) | 43 vs 50 | 0.75 (0.57, 1.00) | 0.92 (0.68, 1.24) |
| 47 | 13 (2) | 54 vs 72 | 0.45 (0.15, 1.36) | 0.59 (0.19, 1.79) | 23 vs 47 | 0.33 (0.09, 1.22) | 0.43 (0.12, 1.59) |
| 48 | 54 (7) | 67 vs 72 | 0.77 (0.43, 1.39) | 0.83 (0.46, 1.51) | 37 vs 48 | 0.65 (0.36, 1.14) | 0.70 (0.39, 1.25) |
| 50 | 5 (1) | 100 vs 72 | NMb | NM | 0 vs 47 | NM | NM |
| 53 | 29 (4) | 62 vs 72 | 0.63 (0.29, 1.36) | 0.93 (0.43, 2.03) | 52 vs 47 | 1.22 (0.58, 2.56) | 1.80 (0.84, 3.84) |
| 54 | 310 (39) | 62 vs 78 | 0.45 (0.33, 0.62) | 0.60 (0.43, 0.84) | 37 vs 53 | 0.53 (0.39, 0.71) | 0.68 (0.49, 0.94) |
| 63 | 577 (73) | 71 vs 75 | 0.78 (0.55, 1.12) | 0.96 (0.66, 1.40) | 46 vs 49 | 0.88 (0.64, 1.20) | 1.07 (0.77, 1.48) |
| 71 | 470 (59) | 69 vs 75 | 0.74 (0.53, 1.01) | 1.21 (0.84, 1.75) | 45 vs 50 | 0.81 (0.61, 1.08) | 1.29 (0.93, 1.79) |
| 73 | 117 (15) | 68 vs 72 | 0.82 (0.54, 1.26) | 0.99 (0.64, 1.53) | 49 vs 47 | 1.09 (0.73, 1.61) | 1.33 (0.89, 1.99) |
| 77 | 182 (32) | 73 vs 71 | 1.07 (0.77, 1.50) | 1.01 (0.72, 1.43) | 52 vs 45 | 1.34 (0.99, 1.81) | 1.31 (0.97, 1.78) |
| 82 | 334 (42) | 62 vs 79 | 0.42 (0.31, 0.58) | 0.52 (0.38, 0.73) | 37 vs 54 | 0.51 (0.38, 0.68) | 0.63 (0.46, 0.85) |
| 84 | 165 (21) | 67 vs 73 | 0.76 (0.52, 1.10) | 0.83 (0.57, 1.21) | 41 vs 49 | 0.72 (0.51, 1.02) | 0.80 (0.56, 1.14) |
| 88 | 35 (4) | 77 vs 72 | 1.34 (0.60, 2.99) | 0.92 (0.40, 2.11) | 54 vs 47 | 1.35 (0.69, 2.68) | 1.02 (0.51, 2.05) |
| 90 | 471 (59) | 72 vs 72 | 1.02 (0.74, 1.39) | 1.29 (0.92, 1.80) | 46 vs 48 | 0.92 (0.69, 1.22) | 1.14 (0.85, 1.55) |
Adjusted odds ratio, odds ratio for mutation after accounting for (other) mutations in the LPV mutation score.
NM, not meaningful.
Virologic response.
The overall proportions of subjects demonstrating at least a 1.0-log10-copies/ml viral load decrease from the baseline or a plasma HIV-1 RNA level of <400 copies/ml at any time point were 72% and 47%, respectively. The virologic responses by the presence or absence of a mutation at each codon are presented for both end points in Table 1. Baseline mutations at codons 10, 20, 24, 36, 54, and 82 were significantly associated with a lowered virologic response in unadjusted analyses by using both end points (Table 1), while the presence of a mutation at codon 30 appeared to be associated with a better virologic response.
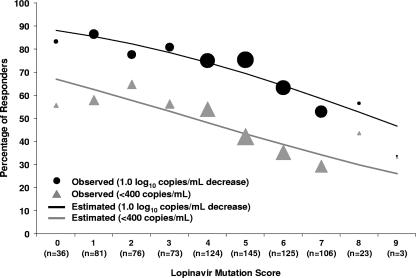
For each virologic end point, a higher LPV mutation score was associated with a decrease in the virologic response by logistic regression (Fig. 1). For example, 79% of the subjects with zero to five mutations demonstrated a 1.0-log10-copies/ml decrease from the baseline in plasma HIV-1 RNA level, whereas 58% of those with six or more mutations experienced such a decrease (P < 0.001, Fisher's exact test). However, some of the 11 mutations within the LPV mutation score were not individually associated with a response (Table 1), suggesting that the score could be improved to provide a better prediction of the virologic response to LPV/r.
FIG. 1.
Observed virologic response rates (circles and triangles) and logistic regression-estimated virologic response rates (lines) in ATU cohort by baseline LPV mutation score. The sizes of the circles and triangles are proportional to the sample size for each category.
Construction of LPV ATU score.
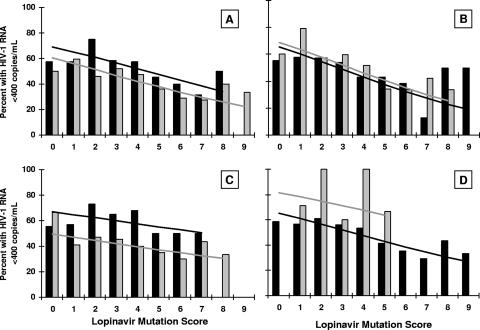
In order to create a score more predictive of a virologic response, the odds ratio for each of the individual 11 codons constituting the LPV mutation score was calculated after adjustment for the contribution of the remaining 10 codons within the score (Table 1). Several codons remained significantly associated with a response (codons 10, 20, 54, and 82; P < 0.05 for both virologic response end points) or demonstrated evidence of a reduced response (codons 24 and 84; odds ratio, <0.85 [see Materials and Methods]). Several codons from the LPV mutation score exhibited no association with the virologic response (codons 53, 63, 71, and 90), and codon 46 demonstrated a trend toward a reduced response but did not meet the criteria to be retained. In a similar manner, the effects of baseline mutations at the remaining 10 codons recorded in the ATU cohort were analyzed after adjustment for the LPV mutation score. By using the criteria described above, the presence of mutations at positions 33, 36, 47, and 48 appeared to be associated with a reduced response. The presence of a substitution at codon 30 was marginally associated with an improved response; whether this represents direct sensitization of the virus to LPV/r or the absence of other key resistance mutations cannot be determined. The remaining codons analyzed were not associated with a virologic response. Graphical representations of the relationships between the absence or the presence of mutations and the probability of achieving plasma HIV-1 RNA levels of <400 copies/ml are displayed in Fig. 2 for a set of representative codons (codons 10, 90, 36, and 30) from the categories discussed above. On the basis of these analyses, a new mutation score, the LPV ATU score, was defined as the number of mutations among the following codons in protease: 10, 20, 24, 33, 36, 47, 48, 54, 82, and 84.
FIG. 2.
Logistic regression analysis of representative codons included or excluded from the LPV ATU score. (A) Codon 10, included in the LPV mutation score and the LPV ATU score; (B) codon 90, included in the LPV mutation score but excluded from the LPV ATU score; (C) codon 36, not included in the LPV mutation score but included in the LPV ATU score; (D) codon 30, not included in either score; the presence of a mutation in this codon was marginally associated with an improved virologic response. Gray and black bars, observed response rates with and without a mutation, respectively, at the codon of interest; gray and black lines, predicted response rate with and without a mutation, respectively, at the codon of interest. The calculation of the LPV mutation score in each panel excludes the codon of interest.
Prediction of virologic response by use of LPV ATU score.
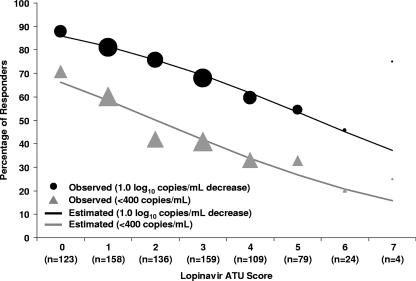
The LPV ATU score was highly associated with a virologic response, with an odds ratio of 0.71 (95% CI, 0.65 to 0.78; P < 0.0001) per additional mutation by using a 1.0-log10 decrease from the baseline in plasma HIV-1 RNA as the end point and an odds ratio of 0.72 (95% CI, 0.65 to 0.79; P < 0.0001) per additional mutation by using a plasma HIV-1 RNA level of <400 copies/ml as the end point (Fig. 3). The proportions of subjects with 1.0-log10 decreases from the baseline in the plasma HIV-1 RNA level and LPV ATU scores of 0 to 2, 3 to 5, or 6 or more were 81%, 62%, and 50%, respectively. The corresponding proportions for subjects achieving plasma HIV-1 RNA levels of <400 copies/ml were 57%, 37%, and 21%, respectively.
FIG. 3.
Observed virologic response rates (circles and triangles) and logistic regression-estimated virologic response rates (lines) in ATU cohort by baseline LPV ATU score. The sizes of the circles and triangles are proportional to the sample size for each category.
Two multivariable analyses were employed to assess the LPV ATU score in the context of other variables. By using stepwise logistic regression analysis, a lower LPV ATU score (odds ratio, 0.80 per additional mutation; P = 0.001) and the previous use of a lower number of PIs (odds ratio, 0.83 per PI; P = 0.012) were associated with a higher likelihood of achieving 1.0-log10 decreases from the baseline in plasma HIV-1 RNA levels. A lower baseline plasma HIV-1 RNA level, the absence of a mutation at codon 82, and the use of a new NNRTI were also associated with this end point. Similar results were observed for the end point of HIV-1 RNA levels of <400 copies/ml.
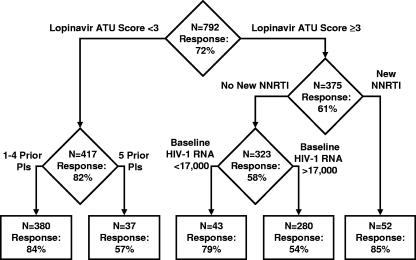
A classification tree for the ATU cohort was developed by using binary recursive partitioning. The final tree contained baseline covariates similar to those in the final stepwise logistic regression model. Notably, the initial partitioning occurred between subjects with lower (zero to two mutations) and higher (three or more mutations) LPV ATU scores. Further partitioning indicated that in subjects with lower LPV ATU scores, the number of PIs used previously was associated with a response, while in subjects with a higher LPV ATU score, the use of a new NNRTI was associated with a response (Fig. 4).
FIG. 4.
Classification tree analysis for assessment of the probability of a 1.0-log10 decrease in the plasma HIV-1 RNA from the baseline.
In a sensitivity analysis, a lower LPV ATU score was statistically significantly associated with a greater maximum decrease from the baseline in the plasma HIV-1 RNA level, with a stronger association than the LPV mutation score either before or after adjustment for other significant variables, such as the number of PIs used previously, the use of a new NNRTI, and the baseline plasma HIV-1 RNA level.
Comparison of LPV ATU score and LPV mutation score in independent populations.
In both of the multivariable analyses of the ATU cohort described above, the LPV ATU score demonstrated a stronger association with a response than the original LPV mutation score. In order to validate this finding in independent data sets, data from three clinical studies of LPV/r-based regimens in PI-experienced subjects were analyzed by using logistic regression with a stepwise selection procedure. In Studies 888 and 957, the LPV ATU score was a statistically significant predictor of a response, while the LPV mutation score did not remain in the model. An apparent breakpoint was observed at between two and three mutations and, to a lesser extent, at between five and six mutations (Table 2). In Study 765, neither scoring method was a significant predictor of a response.
TABLE 2.
Virologic response to LPV/r by LPV ATU score in clinical trials of LPV/r
| LPV ATU score | No. of subjects with virologic response/ total no. of subjects (%)
|
||
|---|---|---|---|
| Study 765 (single PI experienced, NNRTI naive; n = 56) | Study 888 (single PI experienced, NNRTI naive; n = 130) | Study 957 (multiple PI experienced, NNRTI naive; n = 50) | |
| 0 | 14/18 (77.8) | 33/46 (71.7) | 3/4 (75.0) |
| 1 | 13/17 (76.5) | 23/31 (74.2) | 10/10 (100.0) |
| 2 | 7/10 (70.0) | 20/26 (76.9) | 6/6 (100.0) |
| 3 | 8/10 (80.0) | 6/12 (50.0) | 6/9 (66.7) |
| 4 | 0/1 (0.0) | 5/8 (62.5) | 7/11 (63.6) |
| 5 | 2/6 (33.3) | 5/6 (83.3) | |
| 6 | 0/1 (0.0) | 1/2 (50.0) | |
| 7 | 0/2 (0.0) | ||
| 0-2 | 34/45 (76) | 76/103 (74) | 19/20 (95) |
| 3-5 | 8/11 (73) | 13/26 (50) | 18/26 (69) |
| 6 or more | 0/1 (0) | 1/4 (25) | |
| Odds ratio per additional mutation (95% CI) | 0.85 (0.50, 1.44) | 0.77 (0.60, 0.98) | 0.60 (0.39, 0.91) |
| P value | 0.544 | 0.034 | 0.017 |
Finally, a weighted version of the LPV ATU score was evaluated that, based on the findings of Parkin and colleagues (21), assigned threefold weights to mutations at codons 54 and 82. Although it remained unassociated with a response for Study 765, the weighted score had a stronger association with a virologic response than the unweighted score for Studies 888 and 957. For Study 888, the odds ratio for each one-unit increase in the weighted score was 0.83 (95% CI, 0.72 to 0.95), and the P values were 0.005 for the weighted score and 0.034 for the unweighted score. For Study 957, the odds ratio was 0.71 (95% CI, 0.55 to 0.91), and the P values were 0.008 for the weighted score and 0.017 for the unweighted score.
DISCUSSION
We previously derived the LPV mutation score based on the relationship between the genotype of the protease gene and the in vitro susceptibility (phenotype) to LPV of HIV-1 isolates from subjects with virologic failure during PI-based therapy. Although this score was derived from in vitro data, it has proven useful in providing guidance on the clinical use of LPV/r in PI-experienced patients (12, 13). In the present study, we evaluated whether a mutation score based on the virologic response rather than in vitro susceptibility data would provide a superior resistance algorithm. We retrospectively assessed the degree to which mutations at each of 21 codons commonly associated with PI resistance were associated with a virologic response, after accounting for the contribution of the LPV mutation score in the ATU cohort, a large prospective observational study of PI-experienced patients treated with LPV/r-based regimens. Five of the 11 mutations constituting the LPV mutation score were not associated with a virologic response, whereas 4 mutations not contained in the score demonstrated an association with a response. The LPV ATU score was therefore defined as the number of mutations among the following codons in the protease domain: 10, 20, 24, 33, 36, 47, 48, 54, 82, and 84. The LPV ATU score was found to be a better predictor of the virologic response in the ATU cohort than the LPV mutation score, for both an end point based on achievement of a 1.0-log10 decrease in the plasma HIV-1 RNA level and an end point based on achievement of a plasma HIV-1 RNA level of <400 copies/ml. The proportions of subjects with a 1.0-log10 decrease in the plasma HIV-1 RNA level were 81%, 62%, and 50% in those with LPV ATU scores of 0 to 2, 3 to 5, and 6 or more, respectively. The corresponding proportions of subjects achieving plasma HIV-1 RNA levels of <400 copies/ml in each category were 57%, 37%, and 21%, respectively.
In two types of multivariable analyses of the ATU cohort, logistic regression and binary recursive partitioning, the LPV ATU score was found to be a better predictor of a virologic response than the LPV mutation score, even after adjustment for other variables associated with a response, such as the number of PIs used previously, the baseline plasma HIV-1 RNA level, and the use of an NNRTI in NNRTI-naive subjects. These results are not unexpected, since the LPV ATU score was derived from data for the ATU cohort. Therefore, to validate these findings, we compared the performance of the two mutation scores using data for three independent clinical trial populations of PI-experienced subjects and stepwise logistic regression analyses. For the subjects in two of the three studies, both scores were statistically significant predictors of a virologic response, but in both cases, the LPV ATU score was more strongly associated with a response. In each of these two study populations, an apparent “breakpoint” was observed between two and three mutations in the LPV ATU score and, to a lesser extent, between five and six mutations. In the third study, neither score was associated with a response, probably due to the limited degree of baseline LPV resistance in that study, in which only a single subject had a baseline LPV ATU score greater than 3. A version of the LPV ATU score that assigned higher weights to mutations at codons 54 and 82 strengthened the relationship to the response in the first two studies, corroborating the findings of Parkin and colleagues that mutations at these codons are more strongly associated with LPV resistance (21).
The derivation of the LPV mutation score and the LPV ATU score differed in several aspects. The LPV mutation score was derived from univariate analyses of in vitro findings (genotype and phenotype) by using a relatively limited number of isolates (n = 112). In contrast, the LPV ATU score was derived from multivariable analyses of the virologic response and the baseline genotype in a large cohort (n = 792). Together, these differences likely explain why the LPV ATU score is a superior predictor of virologic response, even in independent cohorts.
The predictive value of the LPV ATU score was strengthened further by weighting the mutations at positions 54 and 82 more heavily than the remaining eight mutations, illustrating the weakness of any genotypic score that does not account for the different contributions of amino acid changes to drug resistance. However, a given mutation is likely to have different effects on viral susceptibility, depending on the set of other mutations with which it is present. The optimal weight assigned to any mutation would require more comprehensive analyses, and the use and interpretation of weighted genotypic algorithms would likely be too complicated for routine clinical use. Thus, simple algorithms, such as that used to derive the LPV ATU score, that weigh each mutation equally or that use simple arithmetic multiples (e.g., weights of 3 for positions 54 and 82) may serve as a practical compromise that is useful for estimating drug activity in antiretroviral agent-experienced patients.
Previous studies have shown that reasonable genotypic breakpoints for the LPV mutation score occur between categories of zero to five, six to seven, and eight or more baseline mutations in protease (12). By using the LPV ATU score, the best breakpoints appear to be between categories of zero to two, three to five, and six or more mutations. Lower breakpoints in the LPV ATU score are to be expected, given that several mutations of relatively high prevalence in PI-experienced patients (e.g., at positions 71 and 90) or even antiretroviral agent-naive patients (e.g., at position 63) that are counted in the LPV mutation score are replaced in the LPV ATU score with mutations of relatively low prevalence in PI-experienced patients (e.g., at positions 33, 47, and 48). Correspondingly, the median LPV mutation score and the LPV ATU score for the ATU cohort were 5 and 2, respectively. The different breakpoints between the two scores are therefore not inconsistent, but care must be taken to use the breakpoints that correspond to the correct LPV genotypic algorithm. Finally, these breakpoints are based on the uniformly weighted LPV ATU score (i.e., each mutation is assigned a score of 1) and would potentially change if mutations at positions 54 and 82 were weighted more heavily.
This study has several limitations. First, the genotypic testing for the ATU cohort reported mutations at only the 21 amino acid positions most commonly associated with PI resistance (see Materials and Methods). Thus, any contribution to resistance from mutations at any of the remaining 78 amino acids constituting the protease gene or from any of the protease substrate cleavage sites is not accounted for in the LPV ATU score. Second, although genotypic algorithms derived in the manner described here are valuable for predicting the virologic response to LPV/r in PI-experienced patients, they may not be good predictors of the virologic response to continued LPV/r therapy in patients who experience viral rebound during LPV/r therapy, particularly if they are PI naive. Third, mutations with a prevalence of lower than 1%, such as those at position 50 (n = 5), were deemed too rare for use in statistical analysis. The protease I50V mutation has been shown in other studies to produce significant decreases in susceptibility to LPV (5, 21), as well as to be selected during viral rebound during LPV/r therapy (18). Thus, the LPV ATU score may overestimate the activity of LPV/r in patients with the I50V mutation. Next, while the assessment of our algorithm in independent studies provides corroboration of its utility, it should be noted that our validation cohorts had comparatively small sample sizes. Finally, the ATU cohort was assembled in a period of time when relatively few PIs were available. Analysis of more recent cohorts of patients failing a wider variety of PI-based therapies may allow further refinement of this genotypic algorithm.
In summary, this study provides the basis for a new genotypic algorithm that is useful for assessing the antiviral activities of LPV/r-based regimens in PI-experienced patients infected with drug-resistant HIV-1. The LPV ATU score derived from this analysis was a better predictor of the virologic response than the original LPV mutation score in multiple patient cohorts. This algorithm may be useful in making clinical treatment decisions and in refining genetic and pharmacologic methods for assessing the activity of LPV/r (e.g., genotypic inhibitory quotients [16]).
Acknowledgments
We gratefully acknowledge Barry Bernstein, Scott Brun, Michael Norton, and Debbie Tokimoto (Abbott) for review and discussion of the manuscript; Jeffrey Isaacson (formerly of Abbott) for analysis; and Molly Opferman (M. G. Opferman Consulting) for assistance with the preparation of the manuscript.
Footnotes
Published ahead of print on 18 June 2007.
REFERENCES
- 1.Baxter, J. D., J. M. Schapiro, C. A. Boucher, V. M. Kohlbrenner, D. B. Hall, J. R. Scherer, and D. L. Mayers. 2006. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J. Virol. 80:10794-10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson, C. A., S. G. Deeks, S. C. Brun, R. M. Gulick, J. J. Eron, H. A. Kessler, R. L. Murphy, C. Hicks, M. King, D. Wheeler, J. Feinberg, R. Stryker, P. E. Sax, S. Riddler, M. Thompson, K. Real, A. Hsu, D. Kempf, A. J. Japour, and E. Sun. 2002. Safety and antiviral activity at 48 weeks of lopinavir/ritonavir plus nevirapine and 2 nucleoside reverse-transcriptase inhibitors in human immunodeficiency virus type 1-infected protease inhibitor-experienced patients. J. Infect. Dis. 185:599-607. [DOI] [PubMed] [Google Scholar]
- 3.Brieman, L., J. Friedman, R. Olshen, and C. Stone. 1984. Classification and regression trees. Wadsworth International Group, Belmont, CA.
- 4.Centers for Disease Control and Prevention. 1994. 1994 revised classification system for human immunodeficiency virus infection in children less than 13 years of age. Morb. Mortal. Wkly. Rep. 43(No. RR-12):1-10. [Google Scholar]
- 5.de Mendoza, C., and V. Soriano. 2004. Resistance to HIV protease inhibitors: mechanisms and clinical consequences. Curr. Drug Metab. 5:321-328. [DOI] [PubMed] [Google Scholar]
- 6.Hammer, S. M., M. S. Saag, M. Schechter, J. S. Montaner, R. T. Schooley, D. M. Jacobsen, M. A. Thompson, C. C. Carpenter, M. A. Fischl, B. G. Gazzard, J. M. Gatell, M. S. Hirsch, D. A. Katzenstein, D. D. Richman, S. Vella, P. G. Yeni, and P. A. Volberding. 2006. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society—USA panel. JAMA 296:827-843. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch, M. S., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kuritzkes, R. T. D'Aquila, L. M. Demeter, S. M. Hammer, V. A. Johnson, C. Loveday, J. W. Mellors, D. M. Jacobsen, and D. D. Richman. 2003. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society—USA Panel. Clin. Infect. Dis. 37:113-128. [DOI] [PubMed] [Google Scholar]
- 8.Isaacson, J., D. Kempf, V. Calvez, I. Cohen-Codar, D. Descamps, E. Guillevic, B. Bernstein, E. Sun, J. P. Chauvin, R. Rode, and K. Grebner. 2002. Quantitative estimate of the effect of individual baseline mutations in HIV protease on the virologic response to lopinavir/ritonavir therapy in heavily antiretroviral experienced patients, abstr. 559-T. 9th Conf. Retrovir. Opportunistic Infect., Seattle, WA.
- 9.Johnson, V. A., F. Brun-Vezinet, B. Clotet, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2006. Update of the drug resistance mutations in HIV-1: fall 2006. Top. HIV Med. 14:125-130. [PubMed] [Google Scholar]
- 10.Jung, M., H. Agut, D. Candotti, D. Ingrand, C. Katlama, and J. M. Huraux. 1992. Susceptibility of HIV-1 isolates to zidovudine: correlation between widely applicable culture test and PCR analysis. J. Acquir. Immune Defic. Syndr. 5:359-364. [PubMed] [Google Scholar]
- 11.Kempf, D. J. 2007. Ritonavir and lopinavir/ritonavir. In J. B. Taylor and D. J. Triggle (ed.), Comprehensive medicinal chemistry II, vol. 8. Elsevier, Oxford, United Kingdom.
- 12.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, J. Sylte, B. Richards, B. Bernstein, R. Rode, and E. Sun. 2002. Analysis of the virological response with respect to baseline viral phenotype and genotype in protease inhibitor-experienced HIV-1-infected patients receiving lopinavir/ritonavir therapy. Antivir. Ther. 7:165-174. [PubMed] [Google Scholar]
- 13.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King, M., D. Kempf, J. Isaacson, R. Rode, S. Brun, B. Bernstein, V. Calvez, I. Cohen-Codar, E. Guillevic, J. P. Chauvin, and E. Sun. 2002. Using classification trees to explore relationships between viral genotype and response to lopinavir/ritonavir-based regimens, abstr. 100. XI Int. HIV Drug Resist. Workshop, Seville, Spain.
- 15.Larder, B. A., P. Kellam, and S. D. Kemp. 1991. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS 5:137-144. [DOI] [PubMed] [Google Scholar]
- 16.Marcelin, A. G., I. Cohen-Codar, M. S. King, P. Colson, E. Guillevic, D. Descamps, C. Lamotte, V. Schneider, J. Ritter, M. Segondy, H. Peigue-Lafeuille, L. Morand-Joubert, A. Schmuck, A. Ruffault, P. Palmer, M. L. Chaix, V. Mackiewicz, V. Brodard, J. Izopet, J. Cottalorda, E. Kohli, J. P. Chauvin, D. J. Kempf, G. Peytavin, and V. Calvez. 2005. Virological and pharmacological parameters predicting the response to lopinavir-ritonavir in heavily protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 49:1720-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcelin, A. G., C. Dalban, G. Peytavin, C. Lamotte, R. Agher, C. Delaugerre, M. Wirden, F. Conan, S. Dantin, C. Katlama, D. Costagliola, and V. Calvez. 2004. Clinically relevant interpretation of genotype and relationship to plasma drug concentrations for resistance to saquinavir-ritonavir in human immunodeficiency virus type 1 protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 48:4687-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mo, H., M. S. King, K. King, A. Molla, S. Brun, and D. J. Kempf. 2005. Selection of resistance in protease inhibitor-experienced, human immunodeficiency virus type 1-infected subjects failing lopinavir- and ritonavir-based therapy: mutation patterns and baseline correlates. J. Virol. 79:3329-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nijhuis, M., C. A. Boucher, P. Schipper, T. Leitner, R. Schuurman, and J. Albert. 1998. Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy. Proc. Natl. Acad. Sci. USA 95:14441-14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norton, M., D. Kempf, S. Brun, J. Omachi, P. Cernohous, and M. King. 2004. Genotypic algorithms specific to lopinavir/ritonavir outperform nonspecific lists of PI mutations in predicting virologic response to lopinavir/ritonavir in PI-experienced patients, abstr. 117. XIII Int. HIV Drug Resist. Conf., Tenerife Sur-Costa Adeje, Canary Islands, Spain.
- 21.Parkin, N. T., C. Chappey, and C. J. Petropoulos. 2003. Improving lopinavir genotype algorithm through phenotype correlations: novel mutation patterns and amprenavir cross-resistance. AIDS 17:955-961. [DOI] [PubMed] [Google Scholar]
- 22.Pellegrin, I., D. Breilh, J. M. Ragnaud, S. Boucher, D. Neau, H. Fleury, M. H. Schrive, M. C. Saux, J. L. Pellegrin, E. Lazaro, and M. Vray. 2006. Virological responses to atazanavir-ritonavir-based regimens: resistance-substitutions score and pharmacokinetic parameters (Reyaphar study). Antivir. Ther. 11:421-429. [PubMed] [Google Scholar]
- 23.Pollard, R. B., M. Thompson, C. Hicks, B. Grinsztejn, A. Horban, M. King, M. Norton, P. Cernohous, J. Omachi, and S. Brun. 2004. Phase 3 comparison of lopinavir/ritonavir vs. investigator-selected protease inhibitors in single PI-experienced, NNRTI-naive patients: 48-week results of study M98-888. 7th Congr. Drug Ther. HIV Infect., Glasgow, United Kingdom.
- 24.Vora, S., A. G. Marcelin, H. F. Gunthard, P. Flandre, H. H. Hirsch, B. Masquelier, A. Zinkernagel, G. Peytavin, V. Calvez, L. Perrin, and S. Yerly. 2006. Clinical validation of atazanavir/ritonavir genotypic resistance score in protease inhibitor-experienced patients. AIDS 20:35-40. [DOI] [PubMed] [Google Scholar]
- 25.Wang, D., and B. Larder. 2003. Enhanced prediction of lopinavir resistance from genotype by use of artificial neural networks. J. Infect. Dis. 188:653-660. [DOI] [PubMed] [Google Scholar]