Abstract
Plasmodium malariae, the parasite responsible for quartan malaria, is transmitted in most areas of malaria endemicity and is associated with significant morbidity. The sequence of the gene coding for the enzyme dihydrofolate reductase-thymidylate synthase (DHFR-TS) was obtained from field isolates of P. malariae and from the closely related simian parasite Plasmodium brasilianum. The two sequences were nearly 100% homologous, adding weight to the notion that they represent genetically distinct lines of the same species. A survey of polymorphisms of the dhfr sequences in 35 isolates of P. malariae collected from five countries in Asia and Africa revealed a low number of nonsynonymous mutations in five codons. In five of the isolates collected from southeast Asia, a nonsynonymous mutation was found at one of the three positions known to be associated with antifolate resistance in other Plasmodium species. Five isolates with the wild-type DHFR could be assayed for drug susceptibility in vitro and were found to be sensitive to pyrimethamine (mean 50% inhibitory concentration, 2.24 ng/ml [95% confidence interval, 0.4 to 3.1]).
The emergence and spread of Plasmodium falciparum parasites resistant to chloroquine led to the widespread introduction of the antifol-sulfa combination sulfadoxine-pyrimethamine (SP) for the first-line treatment of malaria. Molecular investigations carried out after the emergence of resistance to SP in P. falciparum led to the identification of dihydropteroate synthase (DHPS) and dihydrofolate reductase (DHFR) as the enzyme targets of sulfadoxine and pyrimethamine, respectively. A series of nonsynonymous point mutations were found to be associated with the resistance phenotype (7, 25, 26, 39). Increasing resistance is caused by the serial acquisition of mutations coding for amino acids in or close to the enzyme binding pocket. Molecular genotyping for resistance markers has become central to assessing and monitoring the prevalence of resistance and thus guiding public health policy.
Chloroquine remains the recommended first-line treatment for Plasmodium vivax infections, but the recent emergence of chloroquine resistance in this parasite (1, 2) is a cause for concern. Clinical studies conducted on the susceptibility of P. vivax lines circulating in Thailand to a variety of antimalarial drugs revealed that the therapeutic responses to SP were poor and associated with high rates of early treatment failure (27, 28). This was unexpected, since although P. falciparum is highly resistant to both chloroquine and SP, chloroquine was, and still is, the first-line treatment for P. vivax infection in Thailand. Subsequent analysis of the Pvdhfr and Pvdhps genes in P. vivax samples collected from Thailand revealed a high frequency of nonsynonymous mutations, particularly those for amino acid residues associated with this high-grade resistance, especially in the Pvdhfr gene (17, 18). Serial acquisition of mutations was associated with increasing resistance in a manner similar to that seen for P. falciparum. The selection of resistance in P. vivax to a drug not generally used for treatment is most likely explained by the high rate of coinfection with P. falciparum and thus exposure to SP (24, 35). These P. vivax parasites, which probably originate from hypnozoites, consistently appear in the bloodstream 3 to 6 weeks after acute treatment of falciparum malaria, when sulfadoxine and pyrimethamine concentrations have reached subinhibitory levels, a situation conducive to the selection of drug-resistant parasites (16).
The sensitive molecular detection techniques that have revealed a high rate of mixed P. falciparum/P. vivax infections have also been used to show that the prevalence of Plasmodium malariae, the causal agent of quartan malaria, is substantially higher than that observed by microscopic examination (21, 30, 32, 34) and that P. malariae is also often found in a mixed infection with P. falciparum. Although hypnozoites have not been demonstrated for this species, P. malariae can persist for decades as a low-grade chronic infection (22, 37). The recommended first-line treatment for P. malariae is chloroquine, and to date only one report from Indonesia has suggested clinical resistance to this drug (23). We wished to ascertain whether there is any evidence that SP pressure might have selected for resistance in P. malariae as it did in P. vivax. Since routine in vitro culturing of P. malariae is not currently available and in vivo studies of drug susceptibility are very difficult because of the low incidence of monoinfection, we sought this evidence by genetic analysis of the gene encoding DHFR. In protozoans, the DHFR protein is linked to a thymidylate synthase (TS) domain forming a bifunctional enzyme encoded by a single gene (15). In this study, the complete coding sequence of the dhfr-ts gene of P. malariae (Pmdhfr-ts) and that of the genetically related parasite of monkeys, Plasmodium brasilianum, were determined. The presence of mutations in the region coding for DHFR was analyzed for isolates from admission blood samples collected from patients infected in Thailand, Lao PDR, Viet Nam, Guinea-Bissau, and Papua New Guinea.
MATERIALS AND METHODS
Plasmodium malariae and Plasmodium brasilianum isolates.
The P. malariae isolates were obtained from samples of either whole blood or dried blood spots (Table 1). Whole-blood samples were collected after informed consent from P. malariae-infected patients admitted to the Bangkok Hospital for Tropical Diseases, Thailand, between 2002 and 2005 (n = 20). Collection was approved by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University. The samples were kept at −20°C until DNA extraction. The dried blood spots containing P. malariae were obtained from four geographic regions: five samples were collected between 2001 and 2004 in Lao PDR, five were collected between 2000 and 2002 in Viet Nam, and two from Papua New Guinea were collected in 2004. For these samples, finger-prick blood was spotted directly onto filter paper (Whatman 3MM chromatography paper), allowed to dry, and stored individually in plastic bags at room temperature until processed. Finally, three samples of P. malariae genomic DNA were purified from isolates collected in Guinea-Bissau in Western Africa in 1995. Genomic DNA from P. brasilianum, purified from a cloned line maintained in Saimiri monkeys, was obtained from MR4 (MRA-349). This New World monkey malaria parasite is considered to be genetically indistinguishable from P. malariae (6).
TABLE 1.
PCR-based detection of Plasmodium species in the samples analyzed
| Sample origin | No. of samples positive for:
|
|||
|---|---|---|---|---|
| P. malariae | P. malariae + P. falciparum | P. malariae + P. vivax | P. malariae + P. falciparum + P. vivax | |
| Thailand | 17 | 1 | 2 | 0 |
| Lao PDR | 5 | 0 | 0 | 0 |
| Viet Nam | 2 | 2 | 0 | 1 |
| Guinea-Bissau | 0 | 3 | 0 | 0 |
| Papua New Guinea | 2 | 0 | 0 | 0 |
| Total | 26 | 6 | 2 | 1 |
Genomic DNA extraction.
DNA was purified from whole-blood samples and dried blood spot samples by use of a commercially available DNA blood kit (QIAGEN, Germany) following the manufacturer's instructions. The volume of DNA solution used as the template for the PCRs was such that 1 μl corresponded to 4 μl of whole blood. The genomic DNA samples were stored at −20°C until use.
Microscopic and PCR analysis of parasite species.
The identification of parasite species was established by microscopic examination of thick and thin blood smears stained with Field's stain. Confirmation of the microscopic diagnosis of P. malariae and detection of other Plasmodium species that might be present in the samples was achieved by nested PCR amplification assays based on the parasite's small-subunit rRNA genes (33, 34). Briefly, genus-specific primers rPLU1 and rPLU5 were used for the primary amplification reaction. Secondary amplifications were performed using the product from the first reaction as the template in separate reactions where another pair of genus-specific primers (rPLU3 and rPLU4) or species-specific primer pairs were used. Samples positive for P. malariae were tested to exclude the presence of Plasmodium knowlesi (19, 31). Control genomic DNAs from each of the four human malaria species, P. knowlesi, and humans were used to confirm the specificity of each primer pair. PCR products from the secondary reactions were amplified only when the genomic DNA of the corresponding species was present, and in each case the fragment sizes corresponded to those predicted.
Oligonucleotide primers.
Degenerate oligonucleotide primers were designed based on conserved regions of the dhfr-ts genes of P. falciparum, of P. vivax, and of the non-human-host species P. berghei, P. chabaudi, and P. gallinaceum (Table 2).
TABLE 2.
PCR primers used for isolation of P. malariae and P. brasilianum dhfr-ts genes
| Primer | Sequence (5′ to 3′)a |
|---|---|
| DHFR-PF | ATGGARSAMSTYTSMGABGTWTTYGA |
| DHFR-PR | TTARGCRGCCATMTCCATDGHDATTTT |
| DHFR-41F | CAAAAYRTYGTRGTNATGGG |
| DHFR-41R | TAAAATTGRCATAAAATATGACAWGG |
| DHFR-42F | ATGGGRARARVHAGYTGGGA |
| DHFR-42R | TTTACATTCCATGCACAYAAAATAATTC |
| DHFR-43F | AAATGYTTYATYATWGGDGG |
| DHFR-43FIV | CCHCCWATRATRAARCATTT |
| Pmdhfr-F | ATGGAGGAAGTCTCAGACGTATTC |
| Pmdhfr-R | TTAGGCGGCCATATCCATAGTTAT |
| Pm5gsp1 | TTGCTGATAGTACTGCTGGTGTT |
| Pm5gsp2 | CTGATGGGGGAGGGATTCTTGGG |
| Pm3gsp1 | GGGAAGCAAATGGAACAAGAGAA |
| Pm3gsp2 | TGGTTTTCAATGGAGGCATTTCG |
| Pm3DVY | CGTCTGTGAGTGATGTGTATACT |
The designation of degenerate sequences at defined positions was made according to IUB nomenclature.
PCR.
Nested or seminested PCR approaches were used in this study to increase the sensitivity of amplification. All amplification reactions were carried out in a total volume of 20 μl in the presence of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 250 nM of each oligonucleotide primer, 125 μM of deoxynucleotide triphosphates, and 4 units of Taq polymerase (Invitrogen). Primary amplification reactions were initiated with 1 μl of the template genomic DNA prepared from whole-blood or dried blood samples, and 1 μl of the product from the primary PCR was used to initiate the secondary amplification. The cycling parameters were as follows: an initial denaturation step at 95°C for 5 min, annealing for 1 min at a temperature defined for different primer pairs (55°C for DHFR-PF plus DHFR-41R, DHFR-PF plus DHFR-43FIV, DHFR-42F plus DHFR-42R, DHFR-41F plus DHFR-PR, DHFR-43F plus Pmdhfr-R, Pmdhfr-F plus Pm5gsp1, Pmdhfr-F plus Pm5gsp2, Pm3DVY plus Pmdhfr-R, Pm3gsp1 plus DHFR-PR, and Pm3gsp2 plus DHFR-PR and 53°C for DHFR-41F plus DHFR-41R and DHFR-PF plus DHFR-42R), extension at 72°C for 2 min, and, finally, denaturation at 94°C for 1 min. After a given number of cycles (30 cycles in primary and 35 cycles in secondary amplification), a final cycle with a 5-min extension step was carried out, and the PCR products were then kept at 4°C until analysis. Negative (no DNA template) and positive control reactions were used in each set of amplifications.
PCR products were detected by electrophoresis of 10 μl of the secondary amplifications in 2% agarose gel (BRL). The DNA was visualized on a UV transilluminator following ethidium bromide staining. The sizes of the amplification products were estimated by comparison to a 100-bp DNA ladder set (New England Biolabs Inc., United Kingdom).
Cloning and sequencing.
Amplification products were either sequenced directly or cloned into the pGEM-T vector (Invitrogen). PCR products and recombinant plasmids were purified using a QIAquick PCR purification kit (QIAGEN, Germany) or a QIAquick Miniprep spin kit (QIAGEN, Germany). DNA sequencing was performed using a DYEnamic ET Dye Terminator cycle sequencing kit and a MegaBACE 500 sequence analyzer (Amersham Bioscience, United Kingdom). The sequencing was performed with both forward and reverse strands from two independent amplification products to confirm that the occurrence of polymorphisms was not due to the presence of a PCR artifact. The sequences have been submitted to GenBank.
Sequence analysis and phylogenetic tree reconstruction.
The DNA sequences obtained from all isolates of P. malariae were analyzed for AT content and codon usage with the sequence manipulation suite program available at http://www.cbio.psu.edu/. The DNA and amino acid sequences were aligned using Clustal W (http://www.ebi.ac.uk/). The tree was constructed using the Neighbor program from the Phylip package, version 3.61 (14).
Antimalarial drug sensitivity assay.
Two milliliters of infected blood was collected in heparinized tubes from patients infected with P. malariae. This blood was centrifuged at 2,500 rpm at 4°C for 5 min. After the removal of plasma and buffy coat, the packed red blood cells were washed three times in RPMI 1640 medium and resuspended to make a 2% cell suspension in the culture medium supplemented with inactivated 10% human AB serum from healthy donors. One-hundred-microliter portions of the cell suspension were added to triplicate wells of microtiter plates predosed with the antimalarial chloroquine, pyrimethamine, or SP (World Health Organization, Manila, The Philippines; quality assurance in 1999). The concentration ranges (2- or 10-fold dilutions) for each drug used in the microtiter plates were 1 to 64 pmol/well for chloroquine, 0.01 to 10,000 pmol/well for pyrimethamine, and 10 to 10,000 pmol/well for SP (80:1). After the erythrocytes were added, the plates were shaken gently to dissolve the drug. The samples were incubated at 37°C in an atmosphere of 5% CO2 for 40 to 70 h depending on the stage of parasite before culture. At the end of incubation, thick and thin blood films were prepared from the samples in each well. Wells without drugs were included as a control. The smears were fixed with methanol and stained with Field's stain before being examined under the microscope. The number of schizonts containing three or more nuclei was counted for the whole thick film. The number of schizonts in the drug-containing wells was compared with the number of schizonts counted in the control wells. Results of each assay at each drug concentration were fitted to a sigmoid curve by use of WinNonlin computer software, version 4.0 (Pharsight Corporation, Mountain View, CA), to determine the 50% inhibitory concentration (IC50).
Nucleotide sequence accession numbers.
Accession numbers of the sequences representative of each P. malariae haplotype that were deposited in GenBank are as follows: for haplotype 1, EF206328, EF206333, and EF206340; for haplotype 2, EF206331; for haplotype 3, EF206334; for haplotype 4, AY846633 to AY846634, EF188271 to EF188273, EF198109 to EF198111, EF206327, EF206330, and EF206335 to EF206336; for haplotype 5, EF206337; for haplotype 6, EF206338; and for haplotype 7, EF206329. Conserved regions of the dhfr-ts genes used to design the primers were from sequences with accession numbers as follows: for P. falciparum, J03772; for P. vivax, X98123; for P. berghei, U12275; for P. chabaudi, M30834; and for P. gallinaceum, AY033582. The P. brasilianum dhfr-ts sequence obtained was assigned accession no. EF188274.
RESULTS
PCR detection of parasite species in the samples.
A total of 35 blood samples reported to harbor P. malariae were obtained from five different geographical locations: Thailand, Lao PDR, Viet Nam, Papua New Guinea, and Guinea-Bissau. For the 20 Thai samples, parasitemia ranged from 2 parasites/thick smear slide to 0.4% parasitemia in the thin film. For the samples from the other areas, parasitemias were less than 0.1%. All 35 samples were diagnosed as being of pure P. malariae infections by microscopy. PCR analysis confirmed the presence of P. malariae and further revealed that mixed infections were present for nine of the samples (Table 1). P. knowlesi was not detected in any of the samples.
Characterization of the DHFR-TS gene from P. malariae and P. brasilianum.
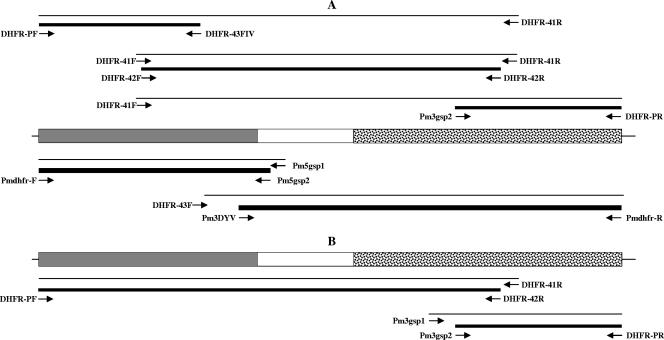
Degenerate oligonucleotide primers designed to generate overlapping amplified fragments corresponding to the dhfr-ts gene were used in amplification reactions using genomic DNA purified from P. malariae (Fig. 1A). Following optimization of the reaction conditions, three overlapping products were obtained, cloned, and sequenced on both strands. The assembled sequence corresponded to the complete Pmdhfr-ts coding sequence, with a size of 1,866 bp. Internal oligonucleotide primer pairs based on the newly acquired sequence were used to confirm that the three fragments were from the same gene (Fig. 1A). The single open reading frame of 1,866 bp encodes a protein of a 622 amino acids (aa) (Fig. 2). The full coding Pmdhfr-ts sequences were obtained for a total of eight isolates, one from Lao PDR (accession no. AY846634) and seven from Thailand (accession no. AY846633, EF188271, EF188272, EF188273, EF198109, EF198110, and EF198111).
FIG. 1.
Schematic representation of the dhfr-ts genes from P. malariae (A) and P. brasilianum (B). The 5′-end region coding for the DHFR domain is shaded in gray, the 3′-end region coding for the TS domain is stippled, and the linker region is left blank. The positions of the oligonucleotides used for nested PCR amplification of bands that were sequenced are indicated. Thin lines indicate the product from the primary amplification reaction, and thick lines indicate the product obtained after the secondary amplification reaction.
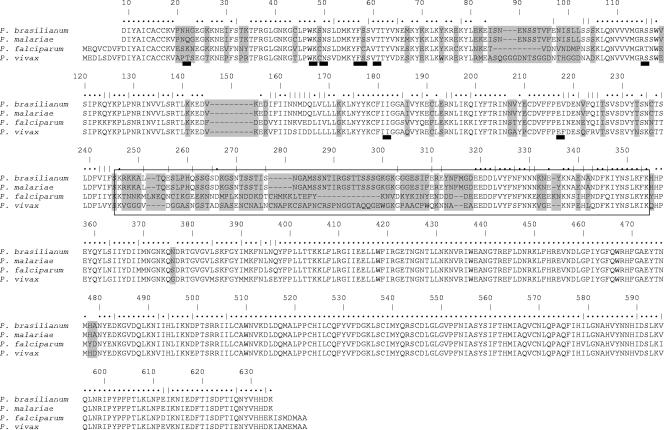
FIG. 2.
Predicted amino acid alignment of the DHFR-TS proteins of P. brasilianum, P. malariae, P. falciparum, and P. vivax. The linker region is in the box. Dots above the sequence denote amino acid identity, and vertical bars denote amino acid similarity. The divergent amino acids are shaded in gray. The thick dark lines below the P. vivax lines indicate the positions of the PmDHFR residues where mutations were observed.
A similar approach was employed using genomic DNA purified from P. brasilianum (Fig. 1B). P. brasilianum is morphologically very similar to P. malariae, both at the erythrocytic and preerythrocytic stages (6), and it is probable that it actually represents a P. malariae population that has adapted to New World monkeys. The sequence obtained, Pbrdhfr-ts (accession no. EF188274), consisted of a single 1,866-bp open reading frame coding for a protein of 622 aa (Fig. 2).
Comparison of dhfr-ts genes across Plasmodium species.
The AT contents of the Pmdhfr-ts and Pbrdhfr-ts genes (69.5%) reflect the intermediate GC content of the P. malariae genome in the same manner that the AT contents of the Pfdhfr-ts (74.7%) and Pvdhfr-ts (52.6%) genes parallel those of the genomes of these two species. The AT compositions of the linker regions for Pmdhfr-ts and Pbrdhfr-ts tended to be lower (62%) than that of the two domains coding for the DHFR and TS enzymes (71%). A similar pattern was noted for the P. vivax gene, in contrast to those of P. falciparum, P. berghei, P. chabaudi, and P. gallinaceum, where the sequences coding for the linker regions tended to have a higher AT content.
Comparison of the complete coding sequences of the eight Pmdhfr-ts genes did not reveal any mutations in the 234-aa DHFR domain or any size variants akin to those observed for the Pvdhfr domain (8-10, 17). Two synonymous substitutions were found in the 101-aa linker, and three nonsynonymous substitutions (D463N, N567S, and N595S) and one synonymous substitution were noted in the 286-aa ts domain. These mutations, though observed only once among the eight sequences, are unlikely to represent artifacts of the amplification reaction, since each sequence was derived from duplicate PCRs. A consensus Pmdhfr-ts predicted peptide sequence, considered to represent the wild-type protein, was then aligned with the corresponding sequences of the seven Plasmodium dhfr-ts genes available at present and with that of Pbrdhfr-ts (Fig. 2). The protein sequence predicted from Pbrdfhr-ts was nearly identical to that derived from Pmdhfr-ts, differing by two residues (Q22H and I206V), both in the dhfr domain. A third synonymous mutation was observed for the Pbrdhfr-ts sequence. The highest degree of amino acid homology between the various species was observed for the ts domain (91% to 95%), which is composed of 286 aa in all the species analyzed. The dhfr domain (size range for the seven species considered, 228 aa to 234 aa) was also relatively conserved, with Pmdhfr showing 71% homology with P. falciparum and P. vivax and 61% to 68% homology with P. berghei, P. chabaudi, and P. gallinaceum. On the other hand, the linker domains vary in size between the species, with that for P. malariae (Pmdhfr-ts) being highly divergent from those for P. falciparum (25%; 94 aa) and P. vivax (8%; 100 aa) and slightly less so from those for the other three species (32% to 34%; 64 aa to 74 aa). It is not known whether the variations in the size or sequence of the linker region have any significant functional consequences.
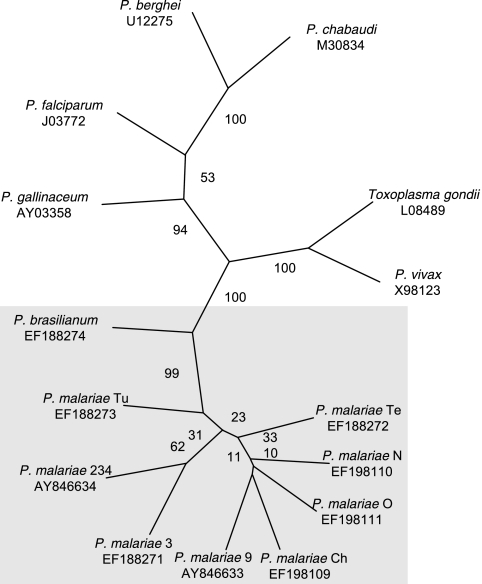
The phylogenetic relationship between Plasmodium species was inferred by neighbor joining and distance (Kimura's two-parameter model). The Toxoplasma gondii dhfr-ts sequence was used as the out-group, because small-subunit rRNA-based phylogenetic analyses of apicomplexan species have placed T. gondii just before the stem that gave rise to Plasmodium (11, 29). The analysis showed that the eight Pmdhfr-ts sequences clustered together along with that of P. brasilianum, as expected (Fig. 3). P. vivax diverged the most from the other Plasmodium spp., P. falciparum was closely related to P. gallinaceum, and two parasites that infect rodents, P. berghei and P. chabaudi, clustered together.
FIG. 3.
Phylogenetic relationship based on the DHFR-TS sequences from eight P. malariae isolates and one P. brasilianum isolate (this article; shaded in gray) and those of five different Plasmodium species (accession numbers for these sequences are provided below each species name).
Mutations in Pmdhfr.
The bulk of the sequence coding for the DHFR domain of P. malariae was amplified from 35 isolates collected from different continents (Table 1). A seminested PCR approach was adopted (Fig. 1A) using P. malariae-specific primer pairs designed to amplify 678 bp of Pmdhfr (excluding the oligonucleotide sequences). The specificity of the reaction was confirmed by the failure to obtain any amplification products when P. falciparum, P. vivax, P. knowlesi, and human genomic DNAs were used as the template. The Pbrdhfr sequence was considered to represent the wild-type sequence, since P. brasilianum could not have been subjected to SP selection pressure in the monkey host. Fourteen distinct mutations were observed at 13 different positions. Seven led to nonsynonymous substitutions at six residues—residues 22, 48, 50, 114 (two different substitutions), 170, and 206—and seven led to synonymous mutations at residues 22, 23, 58, 86, 91, 126, and 148. Six of the seven silent mutations were observed only once, whereas the seventh, at residue 23, was observed for all 35 Pmdhfr sequences. For the nonsynonymous mutations (Table 3), the three at residues 48, 50, and 170 were observed only once, the two affecting residue 114 were observed twice and once, respectively, and those affecting residue 22 were observed for 30 of the 35 sequences, while the mutation observed for residue 206 was found for all 35 Pmdhfr sequences. The equivalent residues for the dhfr genes of P. falciparum and P. vivax are indicated in Table 3, where those previously associated with SP resistance are identified. In total, seven different haplotypes were noted (Table 3).
TABLE 3.
Nonsynonymous mutations observed for P. malariae, P. brasilianum, P. falciparum, and P. vivax
| Organism | Amino acid residuea
|
No. of isolates | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13 | 15 | 22 | 48 | 50 | 57 | 58 | 61 | 114 | 170 | 206 | ||||||||||||||
| P. brasilianum | I | A | H | K | N | F | S | T | S | I | V | 1 | ||||||||||||
| P. malariae | ||||||||||||||||||||||||
| Haplotype 1 | I | A | H | K | N | F | S | T | S | I | I | 3b | ||||||||||||
| Haplotype 2 | I | A | H | K | N | F | S | T | G | I | I | 1 (Viet Nam) | ||||||||||||
| Haplotype 3 | I | A | H | E | N | F | S | T | S | I | I | 1 (Guinea-Bissau) | ||||||||||||
| Haplotype 4 | I | A | Q | K | N | F | S | T | S | I | I | 26c | ||||||||||||
| Haplotype 5 | I | A | Q | K | N | F | S | T | N | I | I | 2 (Thailand) | ||||||||||||
| Haplotype 6 | I | A | Q | K | K | F | S | T | S | I | I | 1 (Lao PDR) | ||||||||||||
| Haplotype 7 | I | A | Q | K | N | F | S | T | S | M | I | 1 (Viet Nam) | ||||||||||||
| P. falciparum | 16 | 51 | 59 | 108 | 164 | |||||||||||||||||||
| A-V | N-I | C-R | S-N/T | I-L | ||||||||||||||||||||
| P. vivax | 13 | 57 | 58 | 61 | 117 | 173 | ||||||||||||||||||
| I-L | F-L/T | S-R | T-M | S-N/T | I-L | |||||||||||||||||||
Residues resulting from nonsynonymous mutations are indicated in boldface.
Two from Viet Nam and one from Guinea-Bissau.
Eighteen from Thailand, 4 from Lao PDR, 2 from Papua New Guinea, 1 from Viet Nam, and 1 from Guinea-Bissau.
In vitro drug sensitivity testing.
In vitro assays of drug sensitivity were successfully carried out with five Thai isolates. For these isolates (starting parasitemias of 0.1% to 0.4%), adequate schizont maturation (three to six merozoites per schizont) was observed for the drug-free control wells, and they could thus be used to evaluate responses to drug treatment. Susceptibilities to chloroquine, pyrimethamine, and SP were evaluated. The inhibition of schizont maturation IC50 values were derived from the sigmoid plots and are summarized in Table 4. These data were compared with those obtained in previous studies with P. falciparum (3, 4, 20) and P. vivax (5). For the P. malariae parasites, schizont maturation was inhibited by all three drugs, particularly pyrimethamine and SP. For these five isolates, the Pmdhfr sequences corresponded to haplotype 4 (Table 3), which differs by a single residue (Q22H) from the wild-type sequence.
TABLE 4.
Susceptibility of in vitro-cultured P. malariae isolates to selected antimalarial drugs
| Drug |
P. malariae
|
P. vivax IC50 (ng/ml) (ref. 5) | P. falciparum IC50 (ng/ml) (ref. 4) | |
|---|---|---|---|---|
| IC50 (ng/ml)a | 95% CIb | |||
| Chloroquine | 74.94 | 64.7-85.2 | 50 | 70 |
| Pyrimethamine | 2.24 | 0.4-3.1 | 8 | 4,083 |
| SP (80:1) | <0.03, 5.71, 6.65c | 10 | ||
The IC50 value is the mean of the values obtained for the five isolates.
CI, confidence interval.
The IC50 could be assessed for only three of the five isolates because of poor slide quality. For one of these isolates, the lowest drug concentration (0.03 ng/ml) of pyrimethamine in the mixture completely inhibited schizont maturation.
DISCUSSION
P. malariae is responsible for quartan malaria. Although infections by P. malariae do not reach high parasitemias and severe clinical symptoms are rarely associated with the acute infection, this species is associated with life-threatening chronic nephropathy in children. P. malariae is globally distributed, but its prevalence was considered to be low and patchy until PCR-based detection methods revealed substantially higher rates of infection, often in mixed infections with P. falciparum (24, 30, 32, 34). It is likely that the contribution of this species to the global malaria health burden is underestimated and that the incidence of P. malariae cases might increase in areas where the successful control of P. falciparum is implemented.
Drug susceptibility studies with P. malariae have been restricted by the paucity of clinical cases and the inability to culture the parasite in vitro. Indications that P. malariae responds more slowly and acquires resistance rapidly to antifolate drugs were provided from experimental infections in humans and from field studies (36, 38). In this study, we have analyzed the sequence of a gene coding for an enzyme, DHFR, associated with resistance to widely used antifolate drugs in 35 P. malariae isolates. The coding region for the DHFR-TS complex was first obtained by amplification using degenerate primers, and this allowed us to design specific primers for the Pmdhfr domain. Given the widespread use of SP and the apparent ease with which P. malariae acquires resistance to pyrimethamine, it was possible that all the sequences obtained might already carry mutations. There are two species, P. brasilianum and P. rodhaini, found in South American and African primates, respectively, that are considered to be P. malariae lines that have adapted to simian hosts. P. brasilianum has been used as an experimental model, and to date partial sequences from three genes of this species, those for the merozoite surface protein 1 (13), the circumsporozoite protein (12), and the small-subunit rRNA (11, 29), were found to be nearly identical to those derived from P. malariae. The dhfr-ts sequence was obtained from P. brasilianum, and this sequence was also found to be nearly identical to that from P. malariae. Taken together with the fact that the two species can infect humans and monkeys, this provides compelling evidence that these parasites are different lines of the same species. Thus, it is reasonable to consider the Pbrdhfr sequence of the laboratory-maintained P. brasilianum line as the wild-type sequence, as it has not been subjected to drug pressure.
The Pmdhfr mutation with the highest frequency (30/35), found in isolates from all geographic locations, occurs at residue 22. This position has not been previously associated with drug resistance in other Plasmodium species, though polymorphisms have been noted at the equivalent positions among isolates of other species (7-9, 17, 18, 39). Five of the parasite isolates for which this Pmdhfr mutation was found could be maintained in culture for a period sufficient to conduct in vitro drug susceptibility assays. The results indicated that parasite development was inhibited at low doses of pyrimethamine and SP (containing sulfadoxine in addition to pyrimethamine). IC50 levels from this study could be compared with our assessment of susceptibility for P. vivax (5), as both P. malariae and P. vivax need folic acid for complete schizont maturation and will not grow in the folate-deficient medium used for P. falciparum sensitivity testing. The IC50 values of SP and pyrimethamine in this study are low, suggesting a full sensitivity of P. malariae to SP and pyrimethamine. It would therefore seem unlikely that the mutation at residue 22 was selected by antifolate drug pressure. For one isolate, a mutation was found at residue 48, a position for which no polymorphisms had been observed for the dhfr genes of other Plasmodium species. The remaining observed mutations led to changes in residues 50, 114, and 170, previously found to be associated with SP resistance in P. falciparum and P. vivax. These single mutations were found in 5 of the 35 isolates, all collected from mainland southeast Asia, i.e., Thailand, Viet Nam, or Lao PDR, where high-level SP resistance is prevalent in P. falciparum and P. vivax. For three isolates, the mutation was found in residue 114, and for two of these it led to the same amino acid substitution (S to N). Mutations at this residue are thought to be the first to be selected when drug pressure is applied on P. falciparum and P. vivax populations (18, 26). Unfortunately, P. malariae isolates with these potential resistance mutations were not available for in vitro testing.
Given that P. malariae is considered to acquire resistance to pyrimethamine rapidly (36, 38), more-frequent mutations in the Pmdhfr gene might have been expected. However, the frequency of resistance-associated mutations observed for Pmdhfr in parasite populations from Thailand and the neighboring Lao PDR were substantially lower than those recorded for the dhfr gene in P. vivax isolates collected in the same region (9, 17, 18). This suggests that these two parasite species have not been subjected to the same selective drug pressure despite, in the case of Thailand, the widespread use of SP as the first-line treatment of P. falciparum from the 1970s until 2001 either as the sole drug or in combination with mefloquine. There are three potential explanations to account for this apparent differential selective pressure. First, the prevalence of P. malariae in Thailand is substantially lower than that of P. vivax. Second, during P. malariae infection episodes, parasite loads rarely exceed 1,000 parasites per μl of blood, whereas for P. vivax, values between 1,000 and 10,000 parasites per μl are common. Thus, the total number of parasites subjected to selective pressure would be lower for P. malariae and the likelihood of selecting a mutant lower. Third, P. vivax parasites emerging from the liver at the beginning of a relapse (consistently at 3 and 6 weeks posttreatment) may be subjected to suboptimal highly selective levels of SP after treatment of a P. falciparum infection episode, whereas P. malariae is not known to produce hypnozoites. Alternatively, mutations in the Pmdhfr gene that confer resistance to antifolate drugs may arise but may be associated with such a high fitness cost that they are selected against and never attain high frequencies in the population.
In conclusion, mutations of the Pmdhfr gene appear to be relatively rare in Thai parasites, implying that the pressure applied when SP was in use did not lead to the widespread selection of resistance in P. malariae, as was the case for P. vivax and P. falciparum. Nonetheless, it would be important to extend this analysis to P. malariae isolates collected from African countries where SP is still in use, or has been until very recently, and where P. malariae prevalence is higher than that found in Thailand. Finally, firm conclusions on any associations between the nonsynonymous mutations found and SP resistance must await further in vivo and in vitro observations as well as kinetic studies on the resulting enzyme isoforms purified as recombinant proteins.
Acknowledgments
We are very grateful to all the patients, the staff, and the nurses at Bangkok Hospital for Tropical Diseases for their help. We also thank the director and staff of the Centre for Malariology, Parasitology and Entomology, Savannakhet Provincial Malaria Station and Phalanxay District Hospital, Vientiane. We thank MR4 for providing us with P. brasilianum DNA contributed by Timothy J. C. Anderson.
This study was part of The Development and Promotion of Science and Technology Talent Project, Thailand, and the Wellcome Trust, Mahidol University, Oxford Tropical Medicine Research Programme funded by the Wellcome Trust of Great Britain. Financial support was provided by a Wellcome Trust fellowship to M.I. (reference no. 066439/2/01/2).
Footnotes
Published ahead of print on 6 August 2007.
REFERENCES
- 1.Baird, J. K., E. Caneta-Miguel, S. Masbar, D. G. Bustos, J. A. Abrenica, A. V. Layawen, J. M. Calulut, B. Leksana, and F. S. Wignall. 1996. Survey of resistance to chloroquine of falciparum and vivax malaria in Palawan, The Philippines. Trans. R. Soc. Trop. Med. Hyg. 90:413-414. [DOI] [PubMed] [Google Scholar]
- 2.Baird, J. K., B. Leksana, S. Masbar, D. J. Fryauff, M. A. Sutanihardja, Suradi, F. S. Wignall, and S. L. Hoffman. 1997. Diagnosis of resistance to chloroquine by Plasmodium vivax: timing of recurrence and whole blood chloroquine levels. Am. J. Trop. Med. Hyg. 56:621-626. [DOI] [PubMed] [Google Scholar]
- 3.Brockman, A., R. N. Price, M. van Vugt, D. G. Heppner, D. Walsh, P. Sookto, T. Wimonwattrawatee, S. Looareesuwan, N. J. White, and F. Nosten. 2000. Plasmodium falciparum antimalarial drug susceptibility on the north-western border of Thailand during five years of extensive use of artesunate-mefloquine. Trans. R. Soc. Trop. Med. Hyg. 94:537-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Childs, G. E., L. Pang, T. Wimonwattrawatee, N. Pooyindee, A. Nanakorn, S. Limchitee, and H. K. Webster. 1987. In vitro mefloquine resistance of Plasmodium falciparum isolated from the Burmese border region of Thailand. Southeast Asian J. Trop. Med. Public Health 18:438-443. [PubMed] [Google Scholar]
- 5.Chotivanich, K., R. Udomsangpetch, W. Chierakul, P. N. Newton, R. Ruangveerayuth, S. Pukrittayakamee, S. Looareesuwan, and N. J. White. 2004. In vitro efficacy of antimalarial drugs against Plasmodium vivax on the western border of Thailand. Am. J. Trop. Med. Hyg. 70:395-397. [PubMed] [Google Scholar]
- 6.Coatney, G. R., W. E. Collins, M. Warren, and P. G. Contacos. 1971. The primate malarias. U.S. Government Printing Office, Washington DC.
- 7.Cowman, A. F., M. J. Morry, B. A. Biggs, G. A. Cross, and S. J. Foote. 1988. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 85:9109-9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Pecoulas, P. E., R. Tahar, T. Ouatas, A. Mazabraud, and L. K. Basco. 1998. Sequence variations in the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene and their relationship with pyrimethamine resistance. Mol. Biochem. Parasitol. 92:265-273. [DOI] [PubMed] [Google Scholar]
- 9.de Pecoulas, P. E., R. Tahar, P. Yi, K. H. Thai, and L. K. Basco. 2004. Genetic variation of the dihydrofolate reductase gene in Plasmodium vivax in Snoul, northeastern Cambodia. Acta Trop. 92:1-6. [DOI] [PubMed] [Google Scholar]
- 10.Eldin de Pecoulas, P., L. K. Basco, R. Tahar, T. Ouatas, and A. Mazabraud. 1998. Analysis of the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene sequence. Gene 211:177-185. [DOI] [PubMed] [Google Scholar]
- 11.Escalante, A. A., and F. J. Ayala. 1994. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc. Natl. Acad. Sci. USA 91:11373-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escalante, A. A., E. Barrio, and F. J. Ayala. 1995. Evolutionary origin of human and primate malarias: evidence from the circumsporozoite protein gene. Mol. Biol. Evol. 12:616-626. [DOI] [PubMed] [Google Scholar]
- 13.Fandeur, T., B. Volney, C. Peneau, and B. de Thoisy. 2000. Monkeys of the rainforest in French Guiana are natural reservoirs for P. brasilianum/P. malariae malaria. Parasitology 120:11-21. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22:521-565. [DOI] [PubMed] [Google Scholar]
- 15.Garrett, C. E., J. A. Coderre, T. D. Meek, E. P. Garvey, D. M. Claman, S. M. Beverley, and D. V. Santi. 1984. A bifunctional thymidylate synthetase-dihydrofolate reductase in protozoa. Mol. Biochem. Parasitol. 11:257-265. [DOI] [PubMed] [Google Scholar]
- 16.Hastings, I. M., W. M. Watkins, and N. J. White. 2002. The evolution of drug-resistant malaria: the role of drug elimination half-life. Philos. Trans. R. Soc. Lond. B 357:505-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imwong, M., S. Pukrittakayamee, S. Looareesuwan, G. Pasvol, J. Poirreiz, N. J. White, and G. Snounou. 2001. Association of genetic mutations in Plasmodium vivax dhfr with resistance to sulfadoxine-pyrimethamine: geographical and clinical correlates. Antimicrob. Agents Chemother. 45:3122-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imwong, M., S. Pukrittayakamee, L. Renia, F. Letourneur, J. P. Charlieu, U. Leartsakulpanich, S. Looareesuwan, N. J. White, and G. Snounou. 2003. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob. Agents Chemother. 47:1514-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jongwutiwes, S., C. Putaporntip, T. Iwasaki, T. Sata, and H. Kanbara. 2004. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg. Infect. Dis. 10:2211-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klinkaewnarong, W., T. Chongsuphajaisiddhi, A. Sabchareon, and C. R. Brockelman. 1985. In vitro sensitivity of Plasmodium falciparum to sulfadoxine and pyrimethamine. Southeast Asian J. Trop. Med. Public Health 16:296-301. [PubMed] [Google Scholar]
- 21.Liu, Q., S. Zhu, S. Mizuno, M. Kimura, P. Liu, S. Isomura, X. Wang, and F. Kawamoto. 1998. Sequence variation in the small-subunit rRNA gene of Plasmodium malariae and prevalence of isolates with the variant sequence in Sichuan, China. J. Clin. Microbiol. 36:3378-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupascu, G., A. Bossie-Agavriloaei, M. Smolinski, E. Ballif Negulici, P. Constantinesco, T. Isfan, D. Petrea, V. Mazilu, and V. Roman. 1963. Le problème des infectionsà P. malariae et les programmes d'éradication du paludisme. Arch. Roum. Pathol. Exp. Microbiol. 22:333-348. [Google Scholar]
- 23.Maguire, J. D., I. W. Sumawinata, S. Masbar, B. Laksana, P. Prodjodipuro, I. Susanti, P. Sismadi, N. Mahmud, M. J. Bangs, and J. K. Baird. 2002. Chloroquine-resistant Plasmodium malariae in south Sumatra, Indonesia. Lancet 360:58-60. [DOI] [PubMed] [Google Scholar]
- 24.Mayxay, M., S. Pukrittayakamee, P. N. Newton, and N. J. White. 2004. Mixed-species malaria infections in humans. Trends Parasitol. 20:233-240. [DOI] [PubMed] [Google Scholar]
- 25.Peterson, D. S., W. K. Milhous, and T. E. Wellems. 1990. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc. Natl. Acad. Sci. USA 87:3018-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plowe, C. V., J. F. Cortese, A. Djimde, O. C. Nwanyanwu, W. M. Watkins, P. A. Winstanley, J. G. Estrada-Franco, R. E. Mollinedo, J. C. Avila, J. L. Cespedes, D. Carter, and O. K. Doumbo. 1997. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J. Infect. Dis. 176:1590-1596. [DOI] [PubMed] [Google Scholar]
- 27.Pukrittayakamee, S., A. Chantra, J. A. Simpson, S. Vanijanonta, R. Clemens, S. Looareesuwan, and N. J. White. 2000. Therapeutic responses to different antimalarial drugs in vivax malaria. Antimicrob. Agents Chemother. 44:1680-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pukrittayakamee, S., M. Imwong, S. Looareesuwan, and N. J. White. 2004. Therapeutic responses to antimalarial and antibacterial drugs in vivax malaria. Acta Trop. 89:351-356. [DOI] [PubMed] [Google Scholar]
- 29.Qari, S. H., Y. P. Shi, N. J. Pieniazek, W. E. Collins, and A. A. Lal. 1996. Phylogenetic relationship among the malaria parasites based on small subunit rRNA gene sequences: monophyletic nature of the human malaria parasite, Plasmodium falciparum. Mol. Phylogenet. Evol. 6:157-165. [DOI] [PubMed] [Google Scholar]
- 30.Scopel, K. K., C. J. Fontes, A. C. Nunes, M. F. Horta, and E. M. Braga. 2004. High prevalence of Plasmodium malariae infections in a Brazilian Amazon endemic area (Apiacas—Mato Grosso State) as detected by polymerase chain reaction. Acta Trop. 90:61-64. [DOI] [PubMed] [Google Scholar]
- 31.Singh, B., L. Kim Sung, A. Matusop, A. Radhakrishnan, S. S. Shamsul, J. Cox-Singh, A. Thomas, and D. J. Conway. 2004. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363:1017-1024. [DOI] [PubMed] [Google Scholar]
- 32.Snounou, G., L. Pinheiro, A. Goncalves, L. Fonseca, F. Dias, K. N. Brown, and V. E. do Rosario. 1993. The importance of sensitive detection of malaria parasites in the human and insect hosts in epidemiological studies, as shown by the analysis of field samples from Guinea Bissau. Trans. R. Soc. Trop. Med. Hyg. 87:649-653. [DOI] [PubMed] [Google Scholar]
- 33.Snounou, G., and B. Singh. 2002. Nested PCR analysis of Plasmodium parasites. Methods Mol. Med. 72:189-203. [DOI] [PubMed] [Google Scholar]
- 34.Snounou, G., S. Viriyakosol, X. P. Zhu, W. Jarra, L. Pinheiro, V. E. do Rosario, S. Thaithong, and K. N. Brown. 1993. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol. Biochem. Parasitol. 61:315-320. [DOI] [PubMed] [Google Scholar]
- 35.Snounou, G., and N. J. White. 2004. The co-existence of Plasmodium: sidelights from falciparum and vivax malaria in Thailand. Trends Parasitol. 20:333-339. [DOI] [PubMed] [Google Scholar]
- 36.Van Goor, W. T., J. G. Lodens, and J. Alier Gomez. 1950. Clinical prophylaxis with proguanil and nivaquine in a community in Java. Doc. Neerl. Indones. Morb. Trop. 2:341-349. [Google Scholar]
- 37.Vinetz, J. M., J. Li, T. F. McCutchan, and D. C. Kaslow. 1998. Plasmodium malariae infection in an asymptomatic 74-year-old Greek woman with splenomegaly. N. Engl. J. Med. 338:367-371. [DOI] [PubMed] [Google Scholar]
- 38.Young, M. D. 1957. Resistance of Plasmodium malariae to pyrimethamine (daraprim). Am. J. Trop. Med. Hyg. 6:621-624. [DOI] [PubMed] [Google Scholar]
- 39.Zolg, J. W., J. R. Plitt, G. X. Chen, and S. Palmer. 1989. Point mutations in the dihydrofolate reductase-thymidylate synthase gene as the molecular basis for pyrimethamine resistance in Plasmodium falciparum. Mol. Biochem. Parasitol. 36:253-262. [DOI] [PubMed] [Google Scholar]