Abstract
By using a high-throughput screening method, a mutant of a uropathogenic Escherichia coli strain affected in the rapA gene was isolated. The mutant formed normal-architecture biofilms but showed decreased penicillin G resistance, although the mutation did not affect planktonic cell resistance. Transcriptome analysis showed that 22 genes were down-regulated in the mutant biofilm. One of these genes was yhcQ, which encodes a putative multidrug resistance pump. Mutants with mutations in this gene also formed biofilms with decreased resistance, although the effect was less pronounced than that of the rapA mutation. Thus, an additional mechanism(s) controlled by a rapA-regulated gene(s) was involved in wild-type biofilm resistance. The search for this mechanism was guided by the fact that another down-regulated gene in rapA biofilms, yeeZ, is suspected to be involved in extra cell wall-related functions. A comparison of the biofilm matrix of the wild-type and rapA strains revealed decreased polysaccharide quantities and coverage in the mutant biofilms. Furthermore, the (fluorescent) functional penicillin G homologue Bocillin FL penetrated the mutant biofilms more readily. The results strongly suggest a dual mechanism for the wild-type biofilm penicillin G resistance, retarded penetration, and effective efflux. The results of studies with an E. coli K-12 strain pointed to the same conclusion. Since efflux and penetration can be general resistance mechanisms, tests were conducted with other antibiotics. The rapA biofilm was also more sensitive to norfloxacin, chloramphenicol, and gentamicin.
Bacterial biofilms are differentiated communities of sessile cells. Infections involving biofilms play a major role in such diverse medical conditions as endocarditis (24), cystic fibrosis (29), otitis media (17), and urinary tract infections (3), presenting a significant threat to human health (12, 16). Although this sessile mode of growth has long been known, the fact that it constitutes a normal part of the bacterial life cycle has been recognized only relatively recently. The medical importance of biofilms lies in the fact that in comparison to their free-living (planktonic) counterparts, biofilms possess increased resistance to antimicrobial agents (12, 16), and consequently, diseases in which biofilms have a dominant role tend to be chronic and difficult to eradicate.
The focus of this study was uropathogenic Escherichia coli (UPEC), which is primarily responsible for urinary tract infections. These affect almost 1 billion women each year. UPEC biofilms can form on the epithelium of the bladder and upper urinary tract (28), as well as intracellularly, evading host defenses (1, 2) and resulting in a disease that is often chronic.
Several genes play a role in biofilm formation (9, 11, 20), but whether the same genes are involved in the development of biofilm resistance has been relatively little studied. Mah et al. (21) identified nvdB gene mutants whose antibiotic resistance was not affected while they were in the planktonic state; furthermore, these mutants were impaired in their biofilm resistance, even though they formed biofilms of normal architecture. Our query in this study was to determine if other genes like nvdB could be identified that accounted for resistance specifically in a biofilm setting and that did so independently of the biofilm architectural features. A high-throughput screening method led to the isolation of a mutant with a mutation in the rapA gene that promised to provide insights into this query. Our results implicate a dual mechanism for enhanced biofilm resistance. To test the generality of the findings, the studies were also extended to an E. coli K-12 strain.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The E. coli strains were maintained in Luria-Bertani (LB) broth. M9 medium supplemented with 0.5% Casamino Acids, 2 mM MgSO4, 25 mM NaCl, and 0.002% FeCl3 (Casamino Acid medium) was used for the flow-cell experiments.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| E. coli strain, plasmid, or primer | Genotype, phenotype,a or sequence | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | supE44 Δlac U169 (φ80 lacZΔM15) hsdR17 recA1 endA1, gyrA96 thi-1 relA1 | Promega |
| K-12 BW25113 | rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | Keio Collection, Japan |
| M1 | E. coli K-12 W3110 ΔrapA, Kmr | Keio Collection, Japan |
| M2 | E. coli K-12 W3110 ΔyhcQ, Kmr | Keio Collection, Japan |
| UPEC | Clinical isolate (AMG1) | Infectious Disease Department, Stanford Medical Center |
| U1 | UPEC but Tn10::rapA; Tcr | This study |
| U2 | U1, pBADrapA; Amr | This study |
| U3 | UPEC, pBADrapAC insertion; Cmr, Amr (rapA deletion strain) | This study |
| U4 | UPEC, pDSRed | This study |
| U5 | U1, pDSRed | This study |
| U6 | UPEC, ΔyhcQ, Kmr | This study |
| Plasmids | ||
| pUC18 | Cloning vector; Amr | NEB, MA |
| pACYC184 | Cloning vector encoding chloramphenicol resistance cassette | NEB, MA |
| pBAD24 | Arabinose-inducible expression vector; Amr | 15 |
| pBADrapA | rapA expression construct; Amr | This study |
| pBADrapAC | pBADrapA with Cmr cassette insertion in rapA gene; Amr, Cmr | This study |
| pDsRed | Constitutively expressed red fluorescent protein vector | Clontech |
| pKD4 | Cloning vector encoding kanamycin resistance cassette | 8 |
| pKD46 | Red recombinase expression plasmid | 8 |
| Primers | ||
| pBADrapAF1 | GAGGAATTCACCATGCCTTTTACACTTGGTCAAb | |
| pBADrapAR1 | TCGGCAAGCTTACTGATGCGTTACAACGATCAAACc | |
| pBADrapACF1 | ACGGGGTACCACGTAAGAGGTTCCAACTTT | |
| pBADrapACR1 | ACGGGTACCGGTGATACCACGATACTATGAC | |
| pkD46yhcQKF1 | TTGTTTTATTTGATATCGCGACTGTTCGTTTGAGGTTGAAGTGTAGGCTGGAGCTGCTTC | |
| pkD46yhcQKR1 | CGAATATGTTGATTAGCAATGGAGAAAATACCCATCGTGACATATGAATATCCTCCTTAG |
Am, ampicillin; Cm, chloramphenicol; Km, kanamycin; Tc, tetracycline.
The underlined sequence is the EcoRI site.
The underlined sequence is the HindIII site.
Mutant isolation.
Transposon mutagenesis was done by using a temperature-sensitive phage, phage λ561, carrying transposon Tn10 (Tcr), as described previously (31, 36). Following incubation at 42°C, which prevents bacteriophage λ replication, transposon insertion mutants were selected on LB-tetracycline plates (15 μg/ml). The mutants were screened for the desired phenotype by comparing their growth inhibition by penicillin with that for the wild type by using the high-throughput method of Das et al. (7). Triplicate wells of microtiter plates were filled with 100- and 200-μl volumes of LB, inoculated with individual strains (initial optical density at 600 nm, 0.06), and grown for 24 h (37°C, no shaking). Following removal of the liquid component of the cultures, the wells were filled with 100- and 200-μl volumes of LB supplemented with 150 μg/ml penicillin G. Twenty-four hours later, bacterial growth was measured with a microtiter plate reader. Das et al. (7) derived an equation correlating the optical densities of the 100- and 200-μl cultures which allows the contribution of biofilm growth to be determined.
MICs for planktonic (MIC) and biofilm (MICadh) cells (defined as the minimum penicillin G concentration that reduced the growth of the planktonic or the biofilm culture by 50% of that observed for the unexposed control, respectively) were determined by the method of Das et al. (7) by using penicillin G concentrations of 0 to 250 μg/ml. The MICs for the UPEC and U1 strains were 50 μg/ml each. The MICadhs for the two strains were 250 and 125 μg/ml, respectively.
Additional methods for biofilm resistance determination.
Three additional methods were used to compare the sensitivities of the wild-type and mutant biofilms to penicillin G and to meet the biofilm biomass need for different experiments: (i) microtiter plate cultivation of biofilms with viability determination by the plate count method, (ii) biofilm cultivation in flow cells followed by staining with BacLight, and (iii) nylon membrane cultivation of biofilms with viability determination by the plate count method. The standard penicillin G treatment regimen was exposure to 350 μg/ml of the antibiotic for 1 h; this protocol has the advantage of being rapid.
Microtiter plate well biofilm cultivation and determination of viability by the plate count method have been described previously (31). The results provided are the averages of at least three independent determinations and are given with error bars of ±1 standard deviation. Cultivation in Plexiglas flow cells was also performed as described previously (31). After 24 h, the biofilms were exposed to penicillin G (in growth medium with flow) for 1 h; in control biofilms, the flow of the medium (without the antibiotic) was continued during this period. Staining with LIVE/DEAD BacLight (Molecular Probes) was used to determine viability. A total of 200 μl of the stain (3 μl of a 1:1 mixture of Syto9 and propidium iodide in 1 ml of saline) was injected with the flow medium into each flow cell. When the probe traversed the length of the flow cell, the flow of the medium was stopped for 5 min to permit exposure of the biofilm to the stain before the flow of the medium was restarted (without the stain) to remove unbound probe. The biofilms were visualized by confocal scanning laser microscopy (CSLM), and images were collected by using consistent CSLM settings (e.g., the laser excitation wavelengths were 476 nm and 543 nm and the emission ranges were 490 to 530 nm and 610 to 650 nm for green and red fluorescence, respectively). The fluorescence intensities of the CSLM images were quantified by using NIH ImageJ freeware (http://rsb.info.nih.gov/ij/index.html). The ratio of the integrated signal intensities from the green fluorescence over the total fluorescence (red plus green fluorescence) was calculated and was used to determine the percent survival compared to that for the untreated controls. In order to reduce nonspecific background light in the images, signal intensity values below a predetermined value were excluded by using the threshold function of the ImageJ software. For all enumeration techniques, with any standard deviation greater than 2.5%, P values were calculated to compare strain differences after treatment by the use of Student's two-tailed t test.
For cultivation on nylon membranes, sterile Hybond nylon membranes (1 by 1 cm) were immersed in LB medium or onto LB plates, the medium or plates were inoculated with an overnight culture of UPEC or strain U1 diluted to an optical density at 600 nm of 0.1, and the medium and the plates were incubated at 37°C. After 24 h of incubation, the liquid culture was discarded and replaced with either fresh LB or fresh LB containing penicillin G, followed by a further 1 h of incubation. For CFU determination, the membranes were removed, rinsed with M9 salt solution, and transferred to tubes containing 10 ml of M9 salt solution. The membranes were then vortexed for 2 min to disperse the biofilms, and the resulting cell suspension was serially diluted and plated in quadruplicate on LB agar plates and incubated overnight.
β-Lactamase assay and PBP profiling.
Assays for β-lactamase were performed as described previously (35). A functional fluorescent penicillin derivative (Bocillin FL; Molecular Probes) was used to determine the penicillin binding protein (PBP) profile of the UPEC and U1 strains as described previously (38). Biofilms grown on the surfaces of deep culture dishes (12 by 6 by 5 cm) were washed once with 100 ml of M9 salt solution and were suspended by scraping the sides of the dishes. The reaction mixture (100 μl) for PBP detection contained the membrane preparation (75 μl; ca. 300 μg protein) and Bocillin FL (33 mg/ml). After incubation in the dark (35°C; 30 min), each reaction mixture (10 μl) was run on a 10% polyacrylamide gel before it was scanned (Typhoon 9400 multiformat imager; excitation wavelength, 488 nm; emission wavelength, 530 nm).
Identification of transposon insertion site.
To identify the gene interrupted by the Tn10 insertion, genomic DNA from strain U1 was isolated and digested with EcoRI, which cleaves Tn10 close to its 3′ end, generating a fragment containing the transposon-derived Tcr marker and the bacterial DNA proximal to the insertion site. Genomic EcoRI fragments were used to generate a clone library in pUC18. This library was electroporated into E. coli DH5α, and clones were selected on 15-μg/ml tetracycline plates. The purified plasmid insert was sequenced at the Stanford University Pan Facility and was identified by using the BLAST (NCBI; http://www.ncbi.nlm.nih.gov/BLAST/) and the WU-BLAST 2.0 (TIGR; http://BLAST.wustl.edu/) programs and the sequences in the GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) and TIGR (http://www.tigr.org/) databases.
Examination of biofilm architecture.
Biofilms were grown in Plexiglas flow cells (31). They were visualized by CSLM with BacLight staining. The biofilm architecture parameters (Table 2) were calculated by using the COMSTAT program (18) coupled to the MATLAB (version 7.2) program (MathWorks, Inc., Natick, MA).
TABLE 2.
Biofilm architecture parameters
| Strain | Thickness (μm) | Roughness coefficient | Surface area/vol |
|---|---|---|---|
| UPEC | 26.3 ± 9.8 | 0.33 ± 0.12 | 2.05 ± 0.02 |
| U1 | 15.1 ± 5.2 | 0.42 ± 0.10 | 2.32 ± 0.08 |
| K-12 BW25113 | 25.5 ± 13.1 | 0.48 ± 0.25 | 2.10 ± 0.75 |
| M1 | 19.7 ± 8.1 | 0.49 ± 0.21 | 2.58 ± 1.34 |
Generation of complementation vector and deletion mutants.
The transposon in the U1 mutant had inserted into the rapA gene. To generate the complementation vector pBADrapA, the rapA gene was amplified by PCR by using the appropriate primers (Table 1). The PCR product was cloned into the arabinose-inducible pBAD24 vector (15). This was electroporated into strain U1, generating strain U2. The induction of RapA protein was confirmed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. A null rapA mutant was generated by replacing a 1.3-kb PvuII fragment of the gene by the chloramphenicol resistance cassette; the latter was amplified from pACYC184. Linearized pBAD rapAC was electroporated into the wild type, and selection for chloramphenicol resistance and ampicillin sensitivity gave the rapA null mutant (strain U3; Table 1). The yhcQ mutant (strain U6) was constructed by the method of Datsenko and Wanner (8) with the specified primers and plasmids (Table 1).
Western blotting of the RapA protein.
Biofilms were grown in deep culture dishes. At the indicated time intervals, 50 ml of planktonic culture from a single dish was harvested, and the remainder was removed by slow vacuum. The biofilms were centrifuged and lysed by using SDS lysis buffer (1% SDS, 20% glycerol, 250 mM Tris-HCl, pH 7.0) and boiling for 5 min. The total protein concentration was determined by the Bio-Rad Dc protein assay, and standardized samples were loaded onto a 12% SDS-polyacrylamide gel. Western blot analysis was performed with a polyclonal RapA antibody as described previously (34).
Transcriptome analysis.
Biofilms were grown for 24 h on Hybond nylon membranes (10 by 10 cm) on LB plates. The biofilms were scraped into 10 ml of Trizol solution (Invitrogen), and the RNA was extracted as described previously (19). Contaminating DNA was removed by DNase I (Ambion) digestion. The RNA samples were further purified with acid phenol-chloroform-isoamyl alcohol (125:24:1; pH 4.5; Ambion) extraction, precipitated with salt-ethanol, and repurified on QIAGEN RNeasy columns. The RNA was quantified spectrophotometrically, and its integrity was checked by gel electrophoresis. cDNA was generated from RNA (10 μg) pooled with internal controls, followed by biotin labeling and processing for microarray analysis. Affymetrix GeneChip E. coli Genome (version 2.0) arrays containing probe sets for the detection of transcripts of UPEC (strain CFT073) were used for transcriptome analysis. Hybridization intensities were quantified by using GeneChip GCOS software (version 1.1.1.052). Background adjustments, scaling, and normalization were performed by using the MAS5 statistical algorithm and spiked-in controls. The data presented are from four independent experiments. The following cutoff values were used to determine changes in expression levels: a P value of <0.01 and a fold change of ≥1.4. Annotations were taken from Affymetrix library files; if a definitive annotation was not found, an analysis with the BLAST program was performed with all bacterial genomes, and analysis of the Pfam database (4) was run to identify any domains.
Binding of penicillin G to biofilms.
Biofilms of each strain were formed for 24 h in 96-well plates as described above. The planktonic culture was removed from 36 wells for each strain, and the biofilms were washed with phosphate-buffered saline (PBS) prior to addition of penicillin solution (200 μl containing 8.3 μg/ml per well, corresponding to 100 μg/ml per 12 wells). This concentration is within the linear range for penicillin determination by the azure C assay (14, 27). The plates were incubated (room temperature, 1 h) prior to removal and pooling of the supernatants from each set of 12 wells. Biofilms from the same set of 12 wells were scraped into PBS (200 μl) and also pooled. The penicillin G concentrations of the supernatant and the biofilm fractions were determined in triplicate by the azure C method by use of a standard curve obtained with 4 to 80 μg of penicillin G. The biofilm biomass in the remaining 60 wells of each plate was determined by crystal violet staining, as described previously (23), and was used to normalize the binding capacity.
Exopolysaccharide staining and analysis.
Biofilms were cultivated in LB medium in six-well Nunc cell culture plates. The exopolysaccharide coverage of 24-h biofilms was determined by ruthenium red staining (26). To analyze the exopolysaccharide composition, the biofilms were harvested, weighed, and resuspended in a volume of PBS corresponding to 16 times their weight. The suspension was vigorously stirred (30 min) and centrifuged (40,000 × g; 2 h). The supernatant was filtered through a 0.22-μm-pore-size filter before it was dialyzed with a Pierce slide-a-lyzer cassette (molecular weight cutoff, 10,000 Da) against 4 liters of distilled H2O (4°C; 36 h; multiple changes of water). The solutions were frozen at −80°C and lyophilized. The dry weight of the exopolysaccharide was normalized to the protein content of the biomass pellet resulting from ultracentrifugation. The protein content was determined by the Bio-Rad Dc assay. Glycosyl composition analysis was performed with the lyophilized exopolysaccharide by combined gas chromatography/mass spectrometry at the Complex Carbohydrate Research Center (University of Georgia).
Penicillin penetration assay.
Red fluorescence protein-expressing UPEC and U1 strains (strains U4 and U5, respectively; Table 1) were grown as biofilms for 24 h in flow cells. Bocillin FL solution (200 μl; final concentration, 33 mg/ml) was added to each flow cell. A high concentration was used to allow saturation of the biofilms with the probe, which permitted the differences in penetration to be determined visually by microscopy. The flow of the medium was stopped once the Bocillin FL traversed the flow cell, and incubation was continued (30 min at 37°C). Unbound probe was removed by restarting the flow of the medium for 5 min. The biofilms were visualized by CSLM, and a series of x-y images from the top to the bottom of each biofilm was collected.
RESULTS
Confirmation that mutant U1 is impaired in resistance to penicillin G only in a biofilm setting.
Spectrophotometric growth measurements obtained by the screening method of Das et al. (7) showed that strain U1 retains the same planktonic penicillin G MIC (MIC, 50 μg/ml) as the wild type but that its biofilm has a decreased MICadh (MICadh, 125 μg/ml versus 250 μg/ml for the wild type). Experiments in shake flask cultures with a range of penicillin G concentrations confirmed that the MIC is 50 μg/ml for both strains.
Multiple test regimens were used to compare the resistance of the strain UPEC and U1 biofilms. The first of these involved cultivation in microtiter plates for 24 h, followed by exposure to penicillin G and enumeration of the CFU. The UPEC biofilm lost 35% viability, and the strain U1 biofilm lost 71% viability (Fig. 1). Untreated control biofilms of the two strains remained fully viable for 24 h, the duration for which the viability tests were conducted. Thus, the greater loss of viability of the U1 biofilm upon penicillin G treatment was not due to its inherent instability.
FIG. 1.
Viable counts after 1 h of penicillin G treatment (350 μg/ml) of 24 h UPEC (solid bars) and U1 (open bars) microtiter biofilms. Control biofilms were grown under identical conditions but were not treated with the antibiotic. Results are averaged from three independent determinations (P < 0.003).
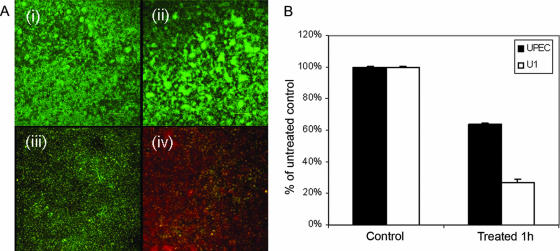
In the second method, biofilms were cultivated on Casamino Acids (rather than LB medium) in Plexiglas flow cells (31) with continuous medium flow and were then treated with penicillin G. Viability was determined by staining with LIVE/DEAD BacLight, which stains viable cells green and dead ones red. As can be seen in Fig. 2A, the UPEC biofilm cells were predominantly green, while those of U1 were red, and the untreated controls of both biofilms were green. Quantification of red and green fluorescence (ImageJ software; NIH freeware) further confirmed the greater loss of viability of the strain U1 biofilms compared to that of the UPEC biofilms (Fig. 2B). Again, it is evident that the greater sensitivity of U1 is not due to the inherent instability of its biofilms.
FIG. 2.
(A) Representative CSLM images (x-y images) of in situ BacLight-stained, flow cell-generated 24-h biofilms: untreated strains UPEC (i) and U1 (ii) incubated for an additional hour and strains UPEC (iii) and U1 (iv) after 1 h of exposure to 350 μg/ml penicillin G. (B) Viability as determined by fluorescence quantification with ImageJ (NIH) software. Magnifications, ×400.
The third method of biofilm cultivation used nylon membranes. Penicillin G treatment of these UPEC biofilms generated no loss of viability but killed 30% of the strain U1 biofilms, a difference which was significant (P < 0.05). Although the results differed somewhat quantitatively, taken together, they confirm the greater sensitivity of the U1 biofilm to penicillin G and show that its sensitivity is independent of the cultivation regimen used.
The biofilms of neither strain possessed β-lactamase activity, nor did they exhibit any differences in their PBP profiles (data not shown), eliminating these factors as causes for their different sensitivities. Architecturally as well, the two flow cell biofilms were very similar, having the typical mushroom-shaped structure (Fig. 3). Analysis with the COMSTAT program indicated other similarities in this respect, although the wild-type biofilm seemed to have a somewhat greater thickness and a decreased surface-to-volume ratio (Table 2).
FIG. 3.
CSLM x-y (A and C) and assembled z-section (B and D) images of strain UPEC (A and B) and strain U1 (C and D) biofilms. Images are representative of the general appearance of each biofilm.
U1 is mutated in the rapA gene.
We determined that the transposon in strain U1 inserted next to nucleotide 655 of the rapA gene. That this generated a loss of a functional nonpolar mutation in this gene is indicated by the following findings: (i) planktonic cultures of U1 exhibited growth inhibition by 0.5 M NaCl, as has been previously reported for a rapA E. coli deletion mutant (33); (ii) U1 planktonic cells containing the arabinose-inducible pBADrapA vector (strain U2; Table 1) showed NaCl growth inhibition, but not when rapA expression was induced by 0.2% arabinose; and (iii) a rapA null mutation constructed in the parent strain (strain U3; Table 1) exhibited a phenotype identical to that of U1 with respect to NaCl and penicillin G sensitivity (data not shown).
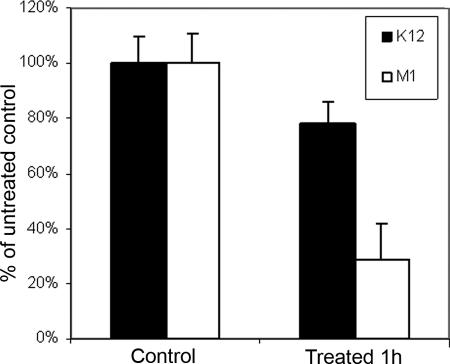
To test whether the effect of the rapA mutation on biofilm sensitivity was confined to the uropathogenic strain of E. coli, we examined the effect of this mutation in another E. coli strain background (a K-12 strain from the Keio Collection; Table 1). This strain and its isogenic rapA deletion mutant (strain M1) were cultivated in Plexiglas flow cells. BacLight staining revealed that the rapA deletion also rendered the biofilm of this strain more sensitive to penicillin G: fluorescence quantification of the images obtained showed 22 and 71% killing of the wild-type and mutant strains, respectively (Fig. 4). Microtiter plate-cultivated biofilms and CFU determination after the antibiotic treatment gave similar results (data not shown).
FIG. 4.
Viability as determined by fluorescence quantification of BacLight-stained images for strain K-12 and M1 biofilms after 1 h of 350-μg/ml penicillin G treatment. Results are averaged from three independent determinations (P < 0.002).
The RapA protein is detected in biofilm cells but not in corresponding planktonic cells.
Western blot analysis showed the presence of the RapA protein at various times (8 to 24 h) in UPEC biofilms but not in biofilm-associated planktonic cells (Fig. 5A). As a control, planktonic cells grown in shake flasks were also analyzed. These contained the protein in early exponential phase (2 and 4 h) but not in later growth stages (Fig. 5B). This is consistent with previous findings (5).
FIG. 5.
Immunoblot analysis at various time points (the numbers above the lanes are in hours) of the RapA protein in UPEC biofilms and associated planktonic cultures (A) and in a UPEC shake flask culture (B).
Transcriptome analysis.
RapA is a homologue of the SWI/SNF superfamily of helicase-like proteins (25, 34), and its role in biofilm resistance was not obvious a priori. Transcriptome analysis was therefore carried out and showed the down-regulation of 22 genes in strain U1 biofilms compared to the gene regulation in UPEC biofilms (Table 3). One of these, the yhcQ gene, encodes a putative multidrug resistance (MDR) pump (22). We therefore compared the penicillin G resistance of the biofilm of a yhcQ deletion mutant (strain U6; Table 1) with those of strain U1 and UPEC in parallel experiments, using microtiter plate wells and CFU measurements. The U6, U1, and UPEC biofilms exhibited 55% ± 2.6%, 71% ± 2.9%, and 35% ± 3.8% killing, respectively, indicating that the loss of the yhcQ gene increases the penicillin G sensitivity of the biofilm but not as much as that of the rapA gene. We also examined the effect of yhcQ mutation in the E. coli K-12 background (strain M2; Table 1). Again, the mutant showed increased penicillin G sensitivity (31% killing, as determined by the quantification of BacLight images), but less than that of the mutant with the rapA mutation (Fig. 4). Flow cell-grown biofilms and BacLight staining gave similar results for both strains U6 and M2 (data not shown). Data for all probe sets are available in Table S1 in the supplemental material.
TABLE 3.
Changes in gene expression between 24-h biofilm cultures of UPEC and U1
| Locus | Gene name | Annotation | Fold change |
|---|---|---|---|
| b0585 | fes | Enterochelin esterase | −3.8 |
| b3242 | yhcR | Hypothetical protein | −3.5 |
| b1705 | ydiE | Hypothetical protein; possible hemin uptake protein | −3.2 |
| b3050 | yqiJ | Putative oxidoreductase | −3.2 |
| b0310 | ykgH | Hypothetical protein; putative membrane protein | −2.9 |
| b1916 | sdiA | Transcriptional regulator of ftsQAZ gene cluster; putative quorum sensing | −2.8 |
| b3071 | yqjI | Hypothetical protein; predicted transcriptional regulator | −2.7 |
| c0394 | NAa | Hypothetical protein | −2.7 |
| c0397 | NA | InsB protein; putative transposase | −2.6 |
| b3241 | yhcQ | Putative membrane protein; putative multidrug efflux pump | −2.4 |
| c1250 | iroN | Siderophore receptor IroN | −2.4 |
| c1624 | NA | Hypothetical protein | −2.1 |
| b2181 | yejG | Hypothetical protein; drug resistance transporter (bicyclomycin) | −2 |
| b0803 | ybiI | Hypothetical protein; putative dnaK suppressor protein | −1.9 |
| b2016 | yeeZ | Nucleoside diphosphate sugar epimerases; cell envelope biogenesis outer membrane/carbohydrate transport and metabolism | −1.9 |
| b1342 | ydaN | Hypothetical protein; possible Mg2+ and Co2+ transporter | −1.7 |
| c2754 | Ada | ADA Regulatory protein; adenosine deaminase regulator protein, regulator of adaptive response | −1.7 |
| b1171 | ymgD | Hypothetical protein | −1.6 |
| b2999 | yghX | Hypothetical protein; possible dienelactone hydrolase | −1.6 |
| b3239 | yhcO | Hypothetical protein; putative barstar-like barnase inhibitor (RNase inhibitor) | −1.6 |
| b3448 | yhhA | Hypothetical protein | −1.6 |
| b1906 | yecH | Hypothetical protein | −1.4 |
NA, not annotated.
Role of reduced polysaccharide coverage of rapA biofilms.
Since rapA mutant biofilms are more sensitive than yhcQ mutant biofilms, a factor(s) in addition to the YhcQ MDR pump controlled by the rapA gene evidently contributes to the wild-type biofilm resistance. The yeeZ gene, putatively concerned with extra-cell wall functions, is also down-regulated in the U1 biofilms (Table 3), and this led us to investigate the matrix-related properties of the two biofilms. The strain U1 biofilms retained twice as much penicillin G as the wild type (Fig. 6). Ruthenium red staining, which specifically stains exopolysaccharide (26), showed that the rapA biofilm stained less intensely than the wild-type biofilm, suggesting reduced exopolysaccharide coverage (Fig. 7). The results of a direct assay were consistent with this finding: while the wild-type biofilm possessed 0.232 ± 0.018 mg polysaccharide per mg of cell protein, the rapA mutant biofilm possessed 0.164 ± 0.017 mg polysaccharide per mg of cell protein. However, the exopolysaccharides of both strains contained similar concentrations of individual sugars (glucose, galactose, rhamnose, mannose, N-acetylfucosamine, N-acetylgalactosamine, N-acetylglucosamine, and xylose) (data not shown).
FIG. 6.
Amount of penicillin G remaining free in solution (black bars) and bound to biofilm (white bars) in 24-h UPEC and U1 biofilms. Penicillin G was quantified by the azure C method.
FIG. 7.
Exopolysaccharide staining of 24-h strain UPEC (i) and strain U1 (ii) biofilms using ruthenium red. The darker red areas represent the increased exopolysaccharide coverage of the biofilm. The images of several biofilms were examined in multiple regions, and representative images are shown.
Did this difference in polysaccharide content affect penicillin G penetration? To answer this question, the green fluorescent functional homologue of penicillin G, Bocillin FL, and UPEC and U1 strains constitutively expressing red fluorescent protein (strains U4 and U5; Table 1) were used to permit visualization by CSLM. Bocillin FL penetrated the biofilms of U1 grown in a flow cell for 24 h more rapidly than the biofilms of UPEC. While in both strains the upper biofilm layers were fully green, in the deeper layers only the U1 biofilms showed this fluorescence (Fig. 8). A video generated from over 100 x-y images taken at 1-μm intervals from the upper to the lower biofilm regions further supports these results (see Video S1 in the supplemental material).
FIG. 8.
Comparable-depth (upper, middle, and bottom) x-y sections through biofilms of equal thickness after in situ exposure to green fluorescently labeled penicillin (Bocillin FL): (A) strain U4 (a UPEC strain constitutively expressing red fluorescent protein) and (B) strain U5 (strain U1 constitutively expressing red fluorescent protein). The presence of green fluorescence indicates the presence of Bocillin FL at the indicated biofilm level. Magnifications, ×400.
Multidrug susceptibility of U1 biofilms.
As MDR pumps, and possibly also the matrix barrier, can confer resistance to multiple drugs, we also compared the sensitivities of the biofilms of the two strains to additional antibiotics. As shown in Fig. 9, the strain U1 biofilm was also more sensitive to norfloxacin; it was found to be some 40% more sensitive to chloramphenicol and gentamicin as well.
FIG. 9.
Survival of strain UPEC (black bars) and strain U1 (white bars) biofilms grown on microtiter plates after exposure for 24 h to a range of norfloxacin concentrations, as determined by CFU count. Percent survival is based on 108 CFU/ml for each strain.
DISCUSSION
A major aim of this study was to identify biofilm features independent of their architecture that account for enhanced resistance. Several cultivation and viability determination methods clearly established that the rapA mutation decreased the penicillin G resistance of E. coli UPEC and K-12 strain biofilms. The cultivation substrata included microtiter plate wells, flow cells, and nylon membranes; and the viability determination methods included spectrophotometry, CFU counts, and BacLight staining. This impairment occurred only in the biofilm setting and did not affect the biofilm architecture, as judged by the evaluation of standard parameters (18). Thus, the rapA mutation promised to provide insights into the basic query of this study. A previous example of such a gene is nvdB, whose absence in Pseudomonas aeruginosa rendered the biofilms much less resistant to several antibiotics, while its absence did not affect planktonic cell resistance or the biofilm architecture (21).
RapA is a homologue of the SWI/SNF superfamily of helicase-like proteins which has been implicated in chromatin remodeling in eukaryotic cells (25, 34). In bacteria, it has never before been implicated in antibiotic resistance. We found that the rapA gene exerted its effect by altering gene regulation in the biofilms, which resulted in lower levels of expression of 22 genes. Of these, one gene was definitively implicated in biofilm resistance, namely, yhcQ: deletion of this gene decreased the biofilm penicillin G resistance of both the E. coli strains. YhcQ is a putative MDR pump (22), and this is clearly a mechanism by which the wild-type E. coli biofilms increase their resistance. It is noteworthy that this pump is also present in the rapA mutant biofilms, but at lower levels, indicating that it is the rapA-mediated increase in its levels that is the critical factor in enhancing biofilm resistance. This role is likely to be specific to the biofilm mode of growth, since the effect of the rapA mutation on resistance is confined to this mode. This is reminiscent of the biofilm-specific role of the MexCD-OprJ pump in the increased resistance of P. aeruginosa biofilms to azithromycin (13), with the difference that the MexCD-OprJ pump is uniquely synthesized in the biofilm setting, while the gene expression levels are altered in the case of the YhcQ pump.
The rapA mutation affected biofilm resistance to a greater extent than the yhcQ mutation, indicating that an additional mechanism(s) controlled by a rapA-regulated gene also has a role in wild-type biofilm resistance. A potential clue to this mechanism was provided by the fact that the yeeZ gene is down-regulated in rapA mutant biofilms. As the putative role of this gene suggests a relationship with extra-cell wall functions (6), the effects of the rapA mutation on matrix-related aspects were compared. The mutant biofilm did indeed exhibit reduced matrix quantity and coverage and showed an expedited penetration of the penicillin G functional homologue, Bocillin FL. This evidence suggests that the wild-type biofilm matrix retards antibiotic penetration and may thus be an additional mechanism of biofilm resistance. While definitive evidence for this involvement remains to be obtained, there are precedents for the role of the matrix in biofilm resistance. For example, the slime of certain staphylococcal strains impedes vancomycin and teicoplanin penetration (30), and Suci et al. (32) showed that the diffusion of ciprofloxacin through P. aeruginosa biofilms is reduced.
As was noted above, P. aeruginosa biofilms also rely on an MDR pump for increased resistance (13), and taken together with the findings of Suci et al. (32), they thus evidently use a dual resistance mechanism involving reduced penetration and active drug efflux (10, 13, 37). Given that we clearly show retarded antibiotic penetration in the wild-type E. coli biofilm, as well as the involvement of the YhcQ pump, it is highly likely that the resistance of the wild-type E. coli biofilm also involves a similar dual resistance mechanism. While one of these mechanisms, namely, the YhcQ pump, most likely has no role in biofilm formation or its structural features, the other mechanism that impaired rapA mutant biofilm resistance is a defective matrix, which is an architectural feature. Consequently, even though the biofilms of strains UPEC and U1 were architecturally similar, as judged by standard gross appearance parameters, it is clear that this did not preclude changes in architectural properties, indicating that gross appearance is not a sufficient criterion for gauging architectural differences.
Both of the mechanisms of resistance controlled by the rapA gene in UPEC biofilms described above can theoretically affect multiple antimicrobials, and indeed, the strain U1 biofilm exhibited greater sensitivity to norfloxacin, chloramphenicol, and gentamicin, which kill bacteria by mechanisms different from that used by penicillin G.
Supplementary Material
Acknowledgments
RapA antibody was a kind gift from D. J. Jin and J. E. Cabrera of NIH, and pDSRed was a kind gift from A. Spormann of Stanford University. We thank Kitty Lee and Jon Mullholland for assistance at the Cell Sciences Imaging Facility at Stanford University.
Carbohydrate analysis was carried out at the University of Georgia Complex Carbohydrate Research Center, supported in part by the U.S. Department of Energy-funded (DE-FG09-93ER-20097) Center for Plant and Microbial Complex Carbohydrates. This work was supported by NASA grant NNA04CC51G (to A.M.), the Office of Biological and Environmental Research of the U.S. Department of Energy under contract DE-AC02-05CH11231 (to G.L.A.), a Stanford Dean's Fellowship (to S.V.L.), NIH training grant T32-AI07328 to the Department of Microbiology and Immunology, and grant no. F32-GM778272 (to M.R.B.) from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 30 July 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Anderson, G. G., K. W. Dodson, T. M. Hooton, and S. J. Hultgren. 2004. Intracellular bacterial communities of uropathogenic Escherichia coli in urinary tract pathogenesis. Trends Microbiol. 12:424-430. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, G. G., S. M. Martin, and S. J. Hultgren. 2004. Host subversion by formation of intracellular bacterial communities in the urinary tract. Microbes Infect. 6:1094-1101. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105-107. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabrera, J. E., and D. J. Jin. 2001. Growth phase and growth rate regulation of the rapA gene, encoding the RNA polymerase-associated protein RapA in Escherichia coli. J. Bacteriol. 183:6126-6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, J., Q. Wang, W. Wang, Y. Wang, L. Wang, and L. Feng. 2006. Characterization of E. coli O24 and O56 O antigen gene clusters reveals a complex evolutionary history of the O24 gene cluster. Curr. Microbiol. 53:470-476. [DOI] [PubMed] [Google Scholar]
- 7.Das, J. R., M. Bhakoo, M. V. Jones, and P. Gilbert. 1998. Changes in the biocide susceptibility of Staphylococcus epidermidis and Escherichia coli cells associated with rapid attachment to plastic surfaces. J. Appl. Microbiol. 84:852-858. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Kievit, T. R., M. D. Parkins, R. J. Gillis, R. Srikumar, H. Ceri, K. Poole, B. H. Iglewski, and D. G. Storey. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 45:1761-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domka, J., J. Lee, T. Bansal, and T. K. Wood. 2007. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9:332-346. [DOI] [PubMed] [Google Scholar]
- 12.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillis, R. J., K. G. White, K. H. Choi, V. E. Wagner, H. P. Schweizer, and B. H. Iglewski. 2005. Molecular basis of azithromycin-resistant Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 49:3858-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gowda, A. T., N. M. Gowda, H. S. Gowda, and K. S. Rangappa. 1985. Application of azure C for the extractive spectrophotometric determination of microgram amounts of penicillin. J. Pharmacol. Methods 13:275-280. [DOI] [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 17.Hall-Stoodley, L., F. Z. Hu, A. Gieseke, L. Nistico, D. Nguyen, J. Hayes, M. Forbes, D. P. Greenberg, B. Dice, A. Burrows, P. A. Wackym, P. Stoodley, J. C. Post, G. D. Ehrlich, and J. E. Kerschner. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296:202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt 10):2395-2407. [DOI] [PubMed] [Google Scholar]
- 19.Hottes, A. K., M. Meewan, D. Yang, N. Arana, P. Romero, H. H. McAdams, and C. Stephens. 2004. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 186:1448-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, Z., F. R. Stirling, and J. Zhu. 2007. Temporal quorum-sensing induction regulates Vibrio cholerae biofilm architecture. Infect. Immun. 75:122-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 22.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 24.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 25.Pazin, M. J., and J. T. Kadonaga. 1997. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell 88:737-740. [DOI] [PubMed] [Google Scholar]
- 26.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahmati, S., S. Yang, A. L. Davidson, and E. L. Zechiedrich. 2002. Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol. Microbiol. 43:677-685. [DOI] [PubMed] [Google Scholar]
- 28.Sakarya, S., G. T. Ertem, S. Oncu, I. Kocak, N. Erol, and S. Oncu. 2003. Escherichia coli bind to urinary bladder epithelium through nonspecific sialic acid mediated adherence. FEMS Immunol. Med. Microbiol. 39:45-50. [DOI] [PubMed] [Google Scholar]
- 29.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 30.Souli, M., and H. Giamarellou. 1998. Effects of slime produced by clinical isolates of coagulase-negative staphylococci on activities of various antimicrobial agents. Antimicrob. Agents Chemother. 42:939-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone, G., P. Wood, L. Dixon, M. Keyhan, and A. Matin. 2002. Tetracycline rapidly reaches all the constituent cells of uropathogenic Escherichia coli biofilms. Antimicrob. Agents Chemother. 46:2458-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suci, P. A., M. W. Mittelman, F. P. Yu, and G. G. Geesey. 1994. Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 38:2125-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sukhodolets, M. V., J. E. Cabrera, H. Zhi, and D. J. Jin. 2001. RapA, a bacterial homolog of SWI2/SNF2, stimulates RNA polymerase recycling in transcription. Genes Dev. 15:3330-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sukhodolets, M. V., and D. J. Jin. 1998. RapA, a novel RNA polymerase-associated protein, is a bacterial homolog of SWI2/SNF2. J. Biol. Chem. 273:7018-7023. [DOI] [PubMed] [Google Scholar]
- 35.Votsch, W., and M. F. Templin. 2000. Characterization of a beta-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and beta-lactamase induction. J. Biol. Chem. 275:39032-39038. [DOI] [PubMed] [Google Scholar]
- 36.Way, J. C., and N. Kleckner. 1984. Essential sites at transposon Tn10 termini. Proc. Natl. Acad. Sci. USA 81:3452-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie, Y., W. X. Jia, W. Zeng, W. Q. Yang, X. Cheng, X. R. Li, L. L. Wang, M. Kang, and Z. R. Zhang. 2005. The action of Pseudomonas aeruginosa biofilms in intrinsic drug resistance. Chin. Med. J. (Engl.) 118:1615-1622. [PubMed] [Google Scholar]
- 38.Zhao, G., T. I. Meier, S. D. Kahl, K. R. Gee, and L. C. Blaszczak. 1999. Bocillin FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 43:1124-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.