Abstract
Pseudomonas aeruginosa is both a model biofilm-forming organism and an opportunistic pathogen responsible for chronic lung infections in cystic fibrosis (CF) patients and infections in burn patients, among other maladies. Here we describe the development of an efficient high-throughput screen to identify small-molecule modulators of biofilm formation. This screen has been run with 66,095 compounds to identify those that prevent biofilm formation without affecting planktonic bacterial growth. The screen is a luminescence-based attachment assay that has been validated with several strains of P. aeruginosa and compared to a well-established but low-throughput crystal violet staining biofilm assay. P. aeruginosa strain PAO1 was selected for use in the screen both because it forms robust biofilms and because genetic information and tools are available for the organism. The attachment-inhibited mutant, strain PAO1 ΔfliC, was used as a screening-positive control. We have also developed and validated a complementary biofilm detachment assay that can be used as an alternative primary screen or secondary screen for the attachment screening-positive compounds. We have determined the potencies of 61 compounds against biofilm attachment and have identified 30 compounds that fall into different structural classes as biofilm attachment inhibitors with 50% effective concentrations of less than 20 μM. These small-molecule inhibitors could lead to the identification of their relevant biofilm targets or potential therapeutics for P. aeruginosa infections.
Establishment of a bacterial infection in the form of a biofilm, a complex, three-dimensional, attached bacterial community, can have devastating consequences for patient morbidity and mortality. Individual cells within a biofilm are slowly growing and are embedded in an exopolymeric substance. These biofilm cells are relatively insensitive to many environmental stresses, including antibiotics and host immune responses (6). Because of the biofilm cells’ intrinsic resistance to antibiotics, the infections that they cause persist, and eradication of these biofilm-related infections is a challenge (7). A new strategy for combating biofilms and persistent infections is desperately needed.
Biofilm infections cause, contribute to, or complicate several conditions, including endocarditis, burns, periodontal disease, ear infections, chronic urinary tract infections, and pneumonia in patients with cystic fibrosis (CF) (7, 29). Devices such as catheters (9) and ventilators (2) that are associated with longer hospital stays and prosthetic and implanted devices such as artificial heart valves, joints, and stents (11) provide surfaces for bacterial attachment, resulting in high rates of morbidity and mortality from nosocomial infections (18, 20). In the United States, these infections are estimated to result in a 20% rate of mortality and to have an annual cost of $1 billion (18), so improvements in the prevention and treatment of biofilm-related persistent infections represent a significant therapeutic opportunity.
In an attempt to identify therapeutic agents for and the therapeutic targets of biofilm-forming opportunistic pathogens, much research has focused on Pseudomonas aeruginosa. In addition to genes for biofilm formation, the P. aeruginosa genome contains genes for several drug efflux pumps, including mexAB-oprM, mexCD-oprJ, and mexXY, that contribute to the organism's ability to resist antibiotics (27) and its ability to create an infection in host tissues, such as the lungs of CF patients (12, 13, 23). P. aeruginosa colonizes the lungs of approximately 21% of CF patients within the first year of life, and by the age of 26 years, nearly 80% of CF patients are colonized. The irreversible damage caused by the recurrent P. aeruginosa lung infections is a serious problem facing most CF patients, and P. aeruginosa contributes to the death of 90% of CF patients (14). Recent advances in antimicrobial therapy and the discovery and use of drugs with antipseudomonal activities, including ceftazidime, aztreonam, ciprofloxacin, and imipenem, have decreased the incidence of P. aeruginosa bacteremia (5). Despite the advances in antibiotics, the incidence of P. aeruginosa bacteremia compared to that of infections caused by other gram-negative bacteria has not drastically declined in the past 20 years (26).
In addition to the medical reasons given above, P. aeruginosa is also an excellent gram-negative bacterial model for the study of the biology of biofilms because of the genetic and physiological information available. In particular, molecular tools which facilitate genetic manipulation have already been developed for P. aeruginosa, particularly strain PAO1. Systematic resources are also available, including microarrays and the P. aeruginosa strain PAO1 genome (36; http://www.pseudomonas.com). Several libraries of P. aeruginosa mutants have been created, and the organisms have been examined for their biofilm-forming phenotype. Mutations in motility, notably, flagellar motility, decreased the amount of initial biofilm attachment (28); and a mutant deficient in flagellum synthesis and initial biofilm attachment (PAO1 ΔfliC) provided an ideal screening-positive phenotype for this work.
A potential strategy for the prevention and treatment of P. aeruginosa biofilm infections would be the use of small molecules to inhibit biofilm development and/or promote biofilm dispersal without the use of a lethal selection pressure. The cells dispersed from a biofilm would be more susceptible to conventional antibiotics and the immune system (8). The halogenated furanones illustrate the potential of small molecules to disrupt bacterial chemical signaling and biofilm formation by some bacteria, although not P. aeruginosa (21). These molecules structurally resemble bacterial acyl-homoserine lactone quorum-sensing molecules (17, 19) and effectively interfere with the reception of the signal, the subsequent gene expression, and the swarming phenotype (24, 25, 33). High-throughput screening (HTS) could be used to identify other compounds effective against P. aeruginosa biofilm development. Ultimately, such compounds could be developed either as small-molecule tools that could be used to study biofilm formation or as therapeutic agents for the prevention and treatment of biofilm infections.
An HTS method requires an appropriate assay. A crystal violet (CV) staining assay had been developed and widely adopted as a means of examining biofilm development on synthetic surfaces (28). CV is a common and inexpensive bacterial cell membrane stain that has been used to quantify biofilm attachment. Although the CV method has been successfully used to screen for attachment and pellicle mutants, it has shortcomings in terms of its dynamic range, the amount of time required per plate, and its reproducibility; and these shortcomings preclude its use as a robust HTS method. An improved assay is needed to screen large, chemically diverse small-molecule libraries in an HTS format.
This report describes the development of luminescence-based biofilm screens for both attachment and detachment and the validation and implementation of the attachment screen to identify small molecules that disrupt biofilm development by P. aeruginosa in an HTS format. The results from the established CV method and the new luminescence-based method are included for comparison; and the development, validation, and use of this protocol to screen 66,095 compounds for their specific antibiofilm activities are described.
MATERIALS AND METHODS
Bacterial strains and media.
The laboratory bacterial strains P. aeruginosa PA14 (31), PAK (3), and PAO1 (22) and clinical isolate P. aeruginosa ZK2870 (16) were used in this study. The commonly studied strain PAO1 was preferred for HTS because a wealth of genetic information and tools are available for this strain. The other commonly used laboratory strains, strains PAK and PA14, were used in this study because considerable information is also available for these strains. Strain ZK2870 was selected because it is a recent clinical isolate that forms robust biofilms on synthetic surfaces. The fliC mutants of PAO1 (15) and PAK (10), which have a delayed biofilm attachment phenotype, were used as positive controls for validation and during experimentation. LB medium was used as the rich medium for starter cultures of P. aeruginosa. The medium used in the biofilm screen consisted of LB at a final concentration of 10% diluted in phosphate-buffered saline (PBS) at pH 7.4 (LB-PBS) (34).
Screening environment.
All robotic liquid handling, compound transfer, and plate reading were performed at the Institute of Chemistry and Cell Biology—Longwood (ICCB-L) at the Harvard Medical School, Boston, MA (http://iccb.med.harvard.edu/). All of the compound libraries that were screened were also maintained by ICCB-L (Table 1). The majority of the libraries and compounds screened were from commercial sources, including ChemBridge (San Diego, CA) and ChemDiv (San Diego, CA), and were selected on the basis of their chemical diversity and availability (http://iccb.med.harvard.edu/screening/compound_libraries/index.htm). A typical screening session involved approximately 10,700 wells (14 plates in duplicate, 4,900 compounds). The compounds were arrayed in 384-well plates at a concentration of 5 mg ml−1 (∼4 to 10 mM) in dimethyl sulfoxide (DMSO).
TABLE 1.
Compound libraries from ICCB-L screened and number of members from each library identified as having activity for preventing biofilm formation by P. aeruginosa PAO1
| Compound group | Library name | No. of compounds | No. of primary screening-positive compoundsa | No. of CPsb | No. of retests from CPs | No. of reordered compounds | No. of compounds with EC50s less than 20 μM |
|---|---|---|---|---|---|---|---|
| Total | 66,095 | 464 | 193 | 83 | 61 | 30 | |
| Known bioactive collection | Biomol ICCB-L Known Bioactives | 480 | 5 | 0 | 0 | 0 | 0 |
| NINDS Custom Collection | 1,040 | 1 | 0 | 0 | 0 | 0 | |
| Prestwick 1 Collection | 1,120 | 1 | 0 | 0 | 0 | 0 | |
| Selected natural | Starr Foundation Extracts 2 | 1,000 | 2 | 1 | 0 | 0 | 0 |
| products | ICBG 1—Fungal Extracts | 851 | 12 | 2 | 0 | 0 | 0 |
| ICBG 2—Fungal Extracts | 460 | 1 | 0 | 0 | 0 | 0 | |
| Organic Fractions—NCI Plant and Fungal Extracts | 1,408 | 8 | 3 | 0 | 0 | 0 | |
| Philippines Plant Extracts 2 | 648 | 13 | 8 | 0 | 0 | 0 | |
| Philippines Plant Extracts 1 | 200 | 1 | 0 | 0 | 0 | 0 | |
| Commercial compounds | ChemDiv 3 | 16,544 | 143 | 61 | 38 | 31 | 13 |
| MixCommercial 5 | 268 | 4 | 1 | 1 | 0 | 0 | |
| Maybridge 4 | 4,576 | 24 | 11 | 3 | 0 | 0 | |
| Biomol-TimTec 1 | 8,166 | 27 | 10 | 6 | 0 | 0 | |
| ChemDiv 2 | 8,560 | 46 | 22 | 10 | 8 | 6 | |
| ChemBridge Microformat | 19,520 | 165 | 70 | 23 | 22 | 11 | |
| ChemDiv Antimitotic Collection | 1,254 | 11 | 4 | 2 | 0 | 0 |
Screening-positive compounds were scored as strong, medium, and weak.
Cherry picks (CPs) were selected from among the screening-positive compounds.
The data were entered into the ICCB-L database and ChemBank (http://chembank.broad.harvard.edu/) and can be compared to the results of other biological assays. The list of screening-positive compounds was compared to the compounds in the database, such that general cytotoxic, nonspecific, and luminescence-inhibiting compounds were eliminated from future consideration. Structure-based groups were assigned by the use of algorithms applied through Pipeline Pilot software (Sci-Tegic).
Biofilm attachment assay protocol.
A schematic representation of the biofilm attachment assay protocol can be found in Fig. 1. Bacterial cultures were archived in glycerol stocks and were stored at −80°C. One day prior to every screening session, fresh aliquots of strains PAO1 and PAO1 ΔfliC were streaked from the archived stock onto a LB plate and the plates were incubated at 37°C. Five milliliters of LB broth in a 14-ml culture tube was inoculated with one colony from each of the plates grown overnight. This culture was incubated at 37°C and was shaken at 250 rpm for 7 h.
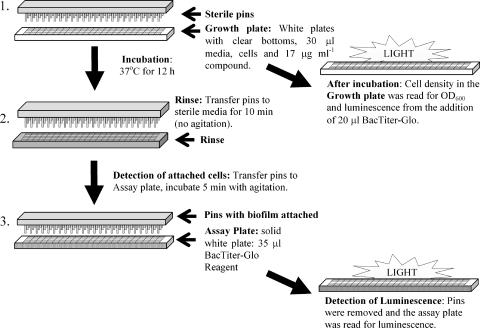
FIG. 1.
Schematic of biofilm attachment assay. Step 1, preparation of the growth plate; step 2, first aspect of plate processing in which the pins are rinsed in the rinse plate and the growth plate is saved for reading by determination of OD600 and luminescence after the addition of the BacTiter-Glo reagent; step 3, determination of biomass attached to the pins by using the BacTiter-Glo reagent and luminescence detection.
Aliquots of LB-PBS were inoculated to a concentration of 1 × 105 CFU ml−1 of strain PAO1 or strain PAO1 ΔfliC liquid culture. The first stage of the assay involved filling each well of a white-walled, clear-bottomed 384-well plate (Costar 3706; Corning, Corning, NY) with 30 μl of medium containing the appropriate culture inoculum; these plates were referred to as the growth plates. For each screening session, two control plates were created, and each experimental plate had on-plate controls, all at 30 μl. Each of the control plates consisted of 128 wells for the negative screening control, which was PAO1; 128 wells for the positive screening control, which was PAO1 ΔfliC; and 128 wells for the no-growth control, which consisted of sterile LB-PBS. Each experimental plate contained 12 wells of the negative screening control, 12 wells of the positive screening control, and 8 wells of the no-growth control. The experimental plates were prepared and assayed in duplicate.
Plate filling, pin transfer, and plate reading took place at ICCB-L. A Well-Mate microfill apparatus (Matrix Technologies, Hudson, NH) was used to dispense 30 μl of strain PAO1-inoculated LB-PBS into the experimental wells. Once they were filled, the plates were gently centrifuged at 168 × g for 1 min by using a Sorvall RT7 Plus and RTH-750 microplate rotor (Thermo Scientific). A total of 100 nl of compound in DMSO (5 mg ml−1) was added to each well by pin transfer by a robot (Epson Robots, Carson, CA) at ICCB-L. Compound from a single stock plate was transferred to duplicate experimental plates by use of the pin transfer device.
Following compound addition, the pin lid tools were inserted into each of the plates and wells to serve as the solid substrate for biofilm attachment (Fig. 1, step 1). The solid substrate for biofilm attachment was an untreated sterile disposable polypropylene 384-pin tool (VP248; V&P Scientific, San Diego, CA). After the pin tools were inserted into the plates, the pin and plate assembly was incubated (without shaking) in a humidified chamber at 37°C for 12 h.
Biofilm attachment assay plate processing and data collection.
The assay procedure is outlined in Fig. 1. This protocol has been developed and validated for P. aeruginosa strains PAO1, PAK, PA14, and ZK2870. Immediately before the plates were removed from the incubator and processed for analysis, rinsed plates, one for each growth plate, were prepared (Fig. 1, step 3). Each well of a clear plastic 384-well plate (Costar 3702) was filled with 35 μl sterile LB-PBS by using the Well-Mate microfill apparatus. The assay reagent, a 25% solution of BacTiter-Glo reagent (Promega, Madison, WI) in PBS, was prepared fresh (55 μl for every well in the experiment). The BacTiter-Glo reagent (Promega) consists of a proprietary combination of a lysis reagent, luciferin, and luciferase and measures the amount of ATP from lysed cells. BacTiter-Glo attachment assay plates (384-well, solid white; Costar 3705) that correspond to each growth plate were prepared, and 35 μl of the 25% BacTiter-Glo reagent was added to each well by using the microfill apparatus.
The incubated plates were carefully removed from the incubator, and in batches of four, the pins were removed and transferred to the respective rinse plate for 10 min without agitation before they were transferred to the attachment assay plates (Fig. 1). The pin tools were exposed to the BacTiter-Glo reagent for 5 min while being agitated twice for 30 s each time, the pin tools were removed, and the luminescence from the attachment assay plate was read by using an Envision plate reader with an ultra-high-sensitivity (UHS) detector (Perkin-Elmer, Waltham, MA). Because the luminescence from the BacTiter-Glo reagent decays linearly (Promega), the time between cell exposure and reading was minimized.
We were interested in identifying compounds that specifically affect biofilm development. Cell growth in the presence of test compounds was also examined by two methods to provide independent assays for cell growth as part of the HTS. After the pins were removed from the clear-bottomed growth plate, the growth plates were centrifuged at 168 × g for 1 min and the absorbance (the optical density at 600 nm [OD600]) was read by using an Envision plate reader.
After the OD600s of the growth plates were analyzed, 20 μl of 25% BacTiter-Glo reagent was added to each well (Fig. 1). The plates were centrifuged at 168 × g for 1 min, and the plates were incubated for 5 min at room temperature in the presence of the reagent before the luminescence was read on an Envision plate reader with an UHS detector.
Biofilm detachment assay protocol.
A detachment assay was used to evaluate the screening-positive compounds (Fig. 2). For the detachment assay, the procedures for the experimental and control plate setup and the addition of the pins for attachment were similar to those for the primary screen; however, for the detachment assay, biofilms were first established on the pins in the absence of compound. After a 12-h incubation to establish the biofilms, the pins were removed from the growth plate, transferred to a rinse plate for 10 min, and then transferred to a detachment plate (Costar 3705) containing 1% LB diluted in PBS (1% LB-PBS) medium and test compounds. The 1% LB-PBS was used in the detachment plate to decrease the growth that occurred during compound incubation but to allow a robust signal from the BacTiter-Glo reagent. The screening controls for the detachment assay included a no-growth control, a screening-negative control used to control for growth, and a screening-positive control consisting of 300 μM EDTA. EDTA was previously shown to aid with the dispersal of established biofilms (1). We observed that 300 μM EDTA was sufficient for detachment without being toxic to the cells.
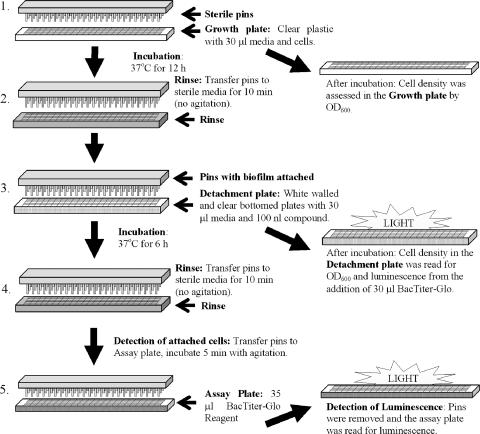
FIG. 2.
Schematic of biofilm detachment assay. Step 1, preparation of the growth plate; step 2, first aspect of plate processing in which the pins are rinsed in the rinse plate and the growth plate is saved for reading by determination of the OD600; step 3, preparation of the detachment plate through the addition of compound and the pins with biofilm attached; after incubation the detachment plate was read at 600 nm and by luminescence after the addition of the BacTiter-Glo reagent; step 4, the pins were rinsed and the pins with the remaining biomass were added to the assay plate for luminescence reading in step 5.
The plate-pin assembly was then incubated for an additional 6 h at 37°C. After incubation in the presence of compound, the pins were transferred to the rinse plate for 10 min and then to a BacTiter-Glo reagent assay plate, and the luminescence was evaluated by using an Envision plate reader with a UHS detector.
The growth plate was examined by measurement of the OD600 by the Envision plate reader. To evaluate the number of cells that detached, 35 μl of 25% BacTiter-Glo reagent was added to the detachment plate, and the luminescence was read as described above. The control to which no compound was added (strain PAO1) was included on each plate and was used to account for growth during the 6-h incubation.
Dose-response curve.
A total of 61 screening-positive compounds were reordered; 39 compounds were from ChemDiv, and 22 compounds were from ChemBridge.
Dose-response curves were determined in triplicate by using the attachment protocol described above. The compounds were dissolved in DMSO to a concentration of 100 mM. Data analysis and 50% effective concentration (EC50) calculations were performed with GraphPad Prism software.
Maintenance protocols.
Before and after the microwell small-bore plate filler manifolds were used (Matrix), they were sterilized by first spraying the outside of the manifold with 70% ethanol and were then rinsed sequentially by priming with 50 ml sterile water, 50 ml 95% ethanol, 50 ml sterile water, and then air. The manifolds were sterilized with an autoclave, as needed. The Envision plate reader protocols were reoptimized weekly.
Assay validation and data analysis.
Assay validation followed the standards determined by the ICCB-L screening facility. In order to quantitatively assess the quality of the HTS assay conditions, the Z′ (Z factor) was determined. This Z′ calculation should not be confused with the z score (standard score) used to standardize individual values (see below) (37). The Z′ calculation takes into consideration both the averages and the standard deviations for the screening-positive control (strain PAO1 ΔfliC or 300 μM EDTA; attachment and detachment assays, respectively) and the screening-negative control (strain PAO1) (37). In order for an assay to be considered robust, the Z′ value should be greater than 0.7 on three consecutive experimental runs of three plates each.
The values from individual wells (compounds) were standardized by using the z score (standard score). The z score was used to demonstrate the units of standard deviation that a value diverges from the mean (37). The z score was ultimately used to rank and score the screening-positive compounds. Optical density and luminescence measurements were both used to determine growth in the presence of the primary screening-positive compounds. The attachment assay was used to evaluate the primary screening-positive compounds; and in this assay, a compound was considered to have a strong, medium, and weak screening-positive result if the z scores for attachment were less than −2.5, −2.5, and −2.0, respectively, and if the levels of growth, as determined by OD600 and luminescence measurements, were greater than −0.1, −0.5, and −0.5, respectively. For the detachment assay, a compound was considered to have detachment activity if the z score for the biofilms remaining attached to the pins was less than −2 and the amount detached into the wells, as measured by determination of the OD600 and luminescence, had a z score greater than −0.5.
To judge the variability and error, each compound was screened in duplicate. To reduce the effects of plate-to-plate variations and day-to-day variations, several controls were incorporated on each assay plate and two designated control plates were included in each screening session. Plate-based controls were incorporated; and assaywide controls were included to determine background levels, signal-to-noise ratios, and standards for normalization.
CV staining assay.
Clear 96- or 384-well plates were inoculated as described above; and additional plates were inoculated with strains PAK, PAK ΔfliC, ZK2870, and PA14. Transferable solid-phase (TSP) pins (Nunc) or VP248 pins were added to the growth plates, and the plates were incubated at 37°C for 12 h. The TSP system was initially used as a proof of principle for the use of pins as a more suitable substrate than well walls for biofilm attachment.
After the 12-h incubation, the pins were removed and rinsed for 10 min in LB-PBS, and the growth plates were analyzed for growth by measuring the absorbance (OD600). The pins were then exposed to 35 μl of 0.1% CV stain for 15 min. After this, the pins were rinsed with water five times for 10 min each time. The CV that sorbed into the cells attached to the pins was then extracted into 95% ethanol and quantified by measuring the OD600.
The attachment of cells to the walls of the growth plates was measured in a similar manner, where 30 μl of CV was added to the 30 μl of culture and the plates were incubated for 15 min. The plate was then rinsed five times for 10 min each time with 60 μl of water; following each rinse, the waste was removed by aspiration. The CV that sorbed to the wall-bound cells was quantified by extraction with 95% ethanol and reading of the OD600.
RESULTS
Development of an HTS assay.
The first step in the development of an HTS assay was to determine if existing assays would be suitable. The CV assay has been well established as a useful method for determining the abilities of different strains to form biofilms on synthetic surfaces. The CV assay has been used reproducibly in 96-well plates (by using the walls of the wells as the substrate). Improvement in the reproducibility of the CV assay in the 96-well format was possible with the use of the TSP system (data not shown). This experiment also demonstrated that biofilms could be grown reproducibly and evaluated by using this removable substrate. We then examined the feasibility of using the CV assay in a 384-well format with VP248 pins.
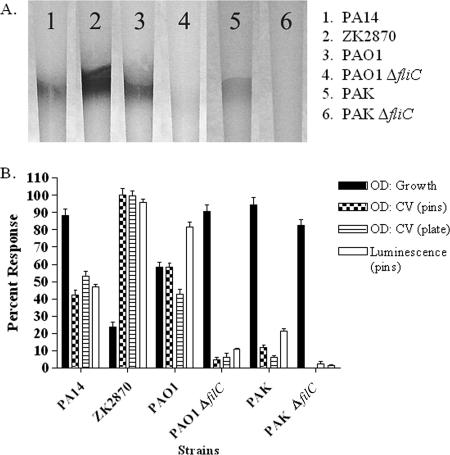
We evaluated the results for several strains of P. aeruginosa that were grown as biofilms on the pins and well walls. The attached biofilms were then subjected to the CV assay, and the results were compared to those obtained by the luminescence-based detection method (Fig. 3). The biofilm attachment of each of the strains to the pins was observed qualitatively (Fig. 3A).
FIG. 3.
Comparison of a luminescence-based HTS assay and the established CV staining methods for biofilm quantification for different strains of P. aeruginosa. (A) Results for CV-stained biofilms on polypropylene VP248 pins; (B) results for CV desorption and luminescence (RLU) as determined for each of the strains tested. The data were normalized (OD for growth plate, 0.12; OD for CV assay with pins, 0.41; OD for CV assay with plates, 1.23; luminescence, 2,100,000) to a percent scale with the background subtracted and presented as the arithmetic mean displayed with the standard deviation. See Table 2 for the Z′ calculations for these assays.
The results of the quantitative comparison of biofilm detection by the luminescence and CV assays are given in Fig. 3B and Table 2. The absorbances of the cell density in the growth plates, CV assays with pins, and CV assay plates with plate walls and the luminescence values were normalized (OD for growth plates, 0.12, OD for CV assays with pins, 0.41; OD for CV assays with plates, 1.23; luminescence assay, 2,100,000) to a percent scale with the background subtracted and are presented as the arithmetic means displayed with the standard deviations in Fig. 3B. Table 2 presents the Z′ values and the dynamic range for each of the assay conditions. The observations from both the CV- and the luminescence-based assays display a dramatic range of biofilm attachment among the strains tested. Strain ZK2870 had the greatest attachment, and strain PAK had the least. The fliC mutants of both PAO1 and PAK attached significantly less (t test, P < 0.001) than their wild-type counterparts. From these results it is evident that the methods give roughly comparable results. However, as shown in Table 2, the reproducibility (Z′ value) was much greater for detection by the luminescence assay than for detection by the CV assay; and the dynamic range for the OD was a maximum of 5.5 times over that for the no-growth control, whereas luminescence had a range of 12,865 times over that for the no-growth control. For these and other reasons—the amount of time per plate and ease of use—a luminescence-based assay was developed for HTS.
TABLE 2.
Calculated Z′ values as a measure of reproducibility of the assays with various strains
| Strain |
Z′a
|
Dynamic rangeb
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OD determination
|
Luminescence assay
|
OD determination
|
Luminescence assay | |||||||||
| Growth plates
|
CV assay with pins
|
CV assay with plates
|
Blank | Mutant control | Growth plates | CV assay with pins | CV assay with plates | |||||
| Blank | Mutant control | Blank | Mutant control | Blank | Mutant control | |||||||
| PA14 | 0.68 | <0.1 | 0.67 | 0.60 | 0.57 | 0.37 | 0.89 | 0.82 | 0.164 (0.007) | 0.268 (0.016) | 0.967 (0.061) | 992,640 (36,261) |
| ZK2870 | <0.1 | 0.53 | 0.80 | 0.77 | 0.77 | 0.70 | 0.92 | 0.89 | 0.085 (0.006) | 0.503 (0.024) | 1.543 (0.063) | 2,006,927 (52,801) |
| PAO1 | 0.60 | <0.1 | 0.77 | 0.75 | 0.47 | 0.32 | 0.86 | 0.82 | 0.127 (0.005) | 0.330 (0.013) | 0.920 (0.057) | 1,715,298 (80,165) |
| PAO1 ΔfliC | 0.68 | NA | <0.1 | NA | <0.1 | NA | 0.87 | NA | 0.165 (0.007) | 0.110 (0.005) | 0.471 (0.043) | 225,330 (9,187) |
| PAK | 0.60 | <0.1 | 0.27 | 0.27 | <0.1 | <0.1 | 0.69 | 0.64 | 0.173 (0.007) | 0.138 (0.008) | 0.269 (0.024) | 436,591 (40,870) |
| PAK ΔfliC | 0.56 | NA | <0.1 | NA | <0.1 | NA | <0.1 | NA | 0.158 (0.006) | 0.085 (0.005) | 0.225 (0.023) | 26,611 (8,569) |
| Blank | NA | NA | NA | NA | NA | NA | NA | NA | 0.057 (0.004) | 0.091 (0.005) | 0.319 (0.031) | 156 (77) |
The mutant control used for the Z′ calculations for strains PA14, ZK2870, and PAO1 was PAO1 ΔfliC, whereas the mutant control used for the Z′ calculation for strain PAK was PAK ΔfliC. NA, calculation is not applicable.
The dynamic range data consist of raw values observed and demonstrate that luminescence had a range of 12,865 times over that for the blank control, whereas the dynamic range for the OD was a maximum of 5.5 times over that for the blank control. Standard deviations are indicated in parentheses.
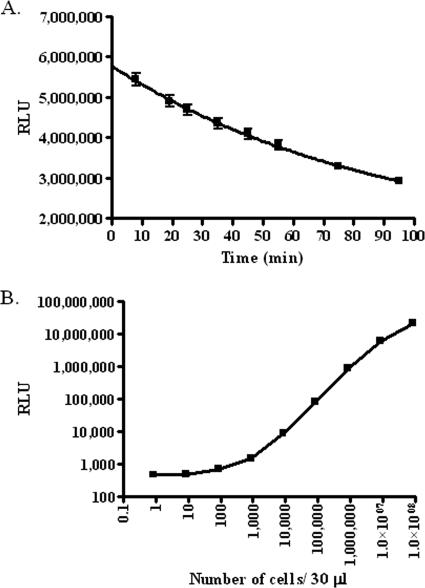
In order to use luminescence, we determined the sensitivity of detection and signal decay using the BacTiter-Glo reagent. The detection of a known number of cells by use of the BacTiter-Glo reagent was examined, and we were able to reliably detect between 8.7 × 102 and 8.7 × 107 cells per well (Fig. 4B). We observed that the luminescence signal (measured as relative light units [RLU]) decays with a half-life of approximately 40 min after the BacTiter-Glo reagent has been added to the cells (Fig. 4A). We examined the lysis efficiency of the BacTiter-Glo reagent with and without sonication and found that there was no added advantage to including a sonication step to either dislodge or lyse the cells.
FIG. 4.
Validation of BacTiter-Glo reagent for use as an HTS tool. The reagent was diluted 1:3 in PBS and was observed for decay under these assay conditions. (A) The half-life of luminescence (RLU) was determined to be approximately 40 min for all cell concentrations tested. The data shown are representative (8.7 × 106 cells). (B) The signal response was also examined for different cell concentrations. The detection limit was between 8.7 × 102 and 8.7 × 107 cells. Both experiments were performed in triplicate.
The luminescence-based assay was validated by calculating Z′ for the biofilm attachment assay for PAO1 versus that for PAO1 ΔfliC of 0.82, a value indicative of a robust HTS assay. The biofilm attachment screen was also validated for strains (versus the results for the control) PAK (versus the results for the PAK ΔfliC), PA14 (versus the results for the PAO1 ΔfliC), and ZK2870 (versus the results for the PAO1 ΔfliC), with the Z′ values calculated to be 0.64, 0.82, and 0.89, respectively. The detachment assay, which we used as a secondary screen to demonstrate the utilities of the compounds for dispersing established biofilms, was validated for PAO1, for which Z′ was calculated to be 0.75, a good value for an HTS assay.
The next step was to define parameters for a biofilm HTS. Initially, several strains were considered for use in the assay. Strain PAO1 was deemed to be the best strain for our assay because it formed consistent and robust biofilms, its genome has been sequenced, and many molecular biology tools are available for the strain. We found that PAO1 biofilms grown in LB-PBS were robust and more manageable and reproducible than biofilms grown in LB at 37°C for 12 h. On the basis of the growth and biofilm attachment curve for PAO1 in LB-PBS (data not shown), it was determined that after a 12-h incubation the cells were in stationary phase and a 12-h incubation was sufficient for robust biofilm formation. We observed that PAO1 can tolerate DMSO concentrations up to 5%, which was well above the concentration usually tested (0.3%).
Implementation of primary screen.
Following validation, the screen was pilot studied with the known bioactive compound collection at ICCB-L. These collections represent 2,640 compounds from three libraries, the Biomol ICCB-L Known Bioactives, the NINDS Custom Collection, and the Prestwick 1 Collection (Table 1). Seven compounds were observed to affect biofilm attachment without affecting growth (three medium screening-positive compounds and four weakly screening-positive compounds), but they were not pursued because they were not potent (Table 1).
Among the collection of 2,640 known bioactive compounds, 50 compounds were found to decrease the growth of P. aeruginosa; and among these were included 29 known antibiotics, including those specifically used to control P. aeruginosa infections, such as cefsulodin sodium (4), pipemidic acid (35), and tobramycin (32).
After the successful screening pilot study, a primary screen was conducted with 66,095 compounds. Table 1 highlights the libraries screened, the number of compounds in each library, the number of attachment screening-positive compounds, and the number of follow-up compounds. In order to be considered a screening-positive compound, several metrics had to be satisfied, all of which were measured by use of the z score. The single attachment metric was measured by determination of luminescence, and the two growth metrics were measured by determination of luminescence and the OD.
In addition to identifying biofilm attachment-inhibiting compounds, we also identified compounds that had general growth-inhibiting characteristics or biofilm-enhancing activities. We observed from the primary screen that 146 compounds inhibited the growth of PAO1, 121 enhanced the growth of PAO1, and 427 increased the biofilm attachment for PAO1. These were not pursued further.
A compound was considered attachment screening positive if it did not cause significant decreases in cell growth and if it significantly decreased attachment in duplicate tests. We identified a total of 464 strong, medium, and weak attachment screening-positive compounds.
We limited our list to 193 (<0.3%) of the most promising compounds for cherry picking (CP). Most of the compounds (n = 182) that we selected for CP were strong screening positive and 11 were medium screening positive. We then removed compounds with poor reproducibility or that were also screening positive by other primary assays performed at ICCB-L, as these compounds may be nonspecific inhibitors for many targets (see Table S2 in the supplemental material). Because the screen was a whole-cell assay with an unknown target, we did not endeavor to develop structure-activity relationships to include or eliminate compounds.
One microliter of each selected CP compound was used to confirm the primary screening results and determine a dose-response. The 193 compounds were retested in triplicate by using the same methods used for the primary screen, with the exception that the 100 nl of compound was diluted in DMSO and the appropriate amount was added to each assay well. Compounds that showed a response similar to that detected in the primary screen were evaluated for a dose-response. Compounds that did not have similar results in the primary screening were retested, and if they still did not have results similar to those of the initial screening, they were considered false positives and were eliminated from future consideration. Using these criteria, we determined the dose-responses of 83 compounds and discarded 110.
By using the remaining 700 nl of the CP aliquot, a single dose-response curve was determined. The initial concentration of compound was 58 μg ml−1, which was then serially diluted 10-fold to 5.8 × 10−13 μg ml−1 (15 points). For the compounds that showed a dose-response, structural clustering was done by using Pipeline Pilot software. At least one representative compound from each of the clusters was purchased for follow-up and confirmation of the dose-response values (Table 3; see Table S1 in the supplemental material).
TABLE 3.
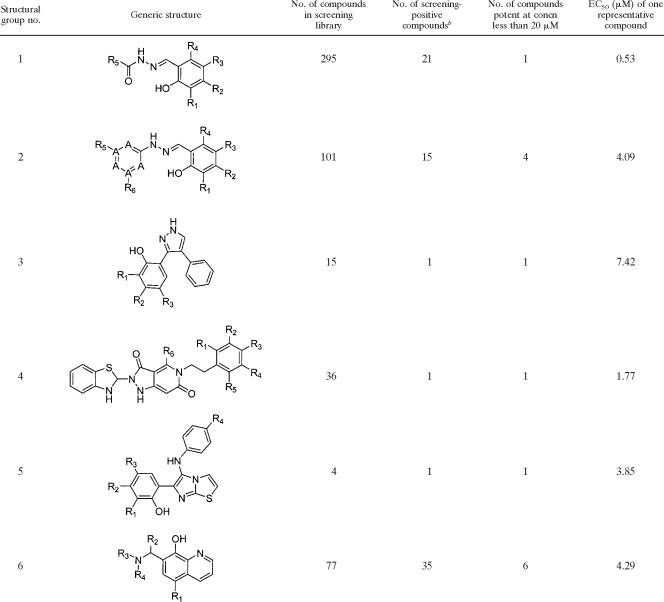
Lead compound structure groups into which 14 of the 30 most potent compounds falla
All 30 structures are available in Table S1 in the supplemental material. The 14 most potent compounds were divided into groups representing structurally related families. The numbers of compounds that also fell into these groups that were in the screening library and that were screening positive are indicated. Six of the groups are represented with generic structures. The notations on molecules R1, R2, R3, etc., indicate points of variation on the molecules. The notation “A” in structural group 2 indicates potential locations of nitrogen atoms on the ring.
b Screening-positive compounds were scored as strong, medium, and weak.
A total of 61 compounds were purchased from either ChemDiv or ChemBridge. The structure and purity of each of the purchased compounds were validated by proton nuclear magnetic resonance imaging by standard methods (30). By using these compounds, a more detailed 20-point dose-response was determined in triplicate for strain PAO1 in both the attachment and the detachment assays. From these reordered compounds, we identified 30 compounds that had EC50s more potent than 20 μM (Table 3). Of the 31 compounds that were not pursued further, all of them were retested at one concentration; while 18 had EC50s greater than 20 μM, it was not possible to determine a dose-response for the other 13. The 30 compounds that showed a potent dose-response in the attachment assay also showed a dose-response in the detachment assay (see Table S1 in the supplemental material).
DISCUSSION
A new therapeutic strategy is desperately needed to combat persistent biofilm infections. In order to identify compounds with antibiofilm activities that could potentially be used as prophylaxis, a robust whole-cell assay was developed. While much research has been done on the biology of biofilms, no one genetic factor has been identified by mutagenesis to effectively and uniformly control biofilm development. A whole-cell approach was desirable because it demonstrated the effectiveness of compounds in a cellular and biofilm community context that examines many relevant targets.
In order to specifically determine the amount of biofilm attached to a surface, we incorporated a pin system because it was inexpensive, disposable, uniform, removable, and easily rinsed and the polypropylene pin surface was suitable for robust biofilm development. By using such a pin system, the difficulties in distinguishing between attached cells, sedimented cells, and cells in suspension were eliminated.
As a comparison of the luminescence assay to the CV based assay for strain PAO1, we observed the luminescence assay to be slightly more robust than the CV assay but much less time-consuming: 11 min per plate for the luminescence assay versus 74 min per plate for the CV assay. Furthermore, the CV assay was messy, and the cost per plate was approximately the same as that of the luminescence assay. In addition, the CV method did not lend itself to easy screening for both planktonic and biofilm growth. We found that although the CV method has been successfully used to screen for attachment and pellicle mutants, it has shortcomings that prohibit it from becoming a robust HTS assay.
The luminescence assay was used to identify 30 compounds that fell into six structural classes as biofilm attachment inhibitors with EC50s of less than 20 μM. These compounds and the classes that they represent do not have known bacterial targets or known modes of action (Table 3; see also Table S1 in the supplemental material). The compounds inhibit biofilm development at concentrations well below their growth-inhibitory levels and are effective at preventing biofilm development by P. aeruginosa. All of the compounds identified were selective for inhibiting biofilm development without affecting growth.
We have also developed an HTS assay for detachment. This assay was used to demonstrate the activities of biofilm attachment-inhibiting compounds at disrupting established biofilms. The biofilm detachment screen could also be used as an HTS assay to identify compounds which act specifically by dispersing established biofilms. The results of such a detachment assay would be clinically relevant to established infections. However, we deemed the attachment assay to be more desirable for a primary screen due to time-per-assay considerations and material costs. We used the detachment assay as a secondary assay to demonstrate the potential efficacies of these compounds for disrupting established biofilms (see Table S1 in the supplemental material).
Although some of the chemical structures suggest that the hit compounds may be metal chelators, the relevance of this to the assay results and the metals being chelated remains unknown. We can report that less than 7 to 15% of the total compounds screened for each compound class were hits, and this likely indicates that these compounds have specific activity other than a general chelating effect. Future work will focus on the physiological effects of these compounds and their use in screens of multiple P. aeruginosa strains.
Supplementary Material
Acknowledgments
We thank the ICCB-L (Harvard Medical School) screening staff for their technical support: Caroline Shamu, Stewart Rudnicki, Katrina Schulberg, David Fletcher, David Wrobel, and Sean Johnston. We also thank Kyungae Lee (NERCE, Harvard Medical School).
We acknowledge the Cystic Fibrosis Foundation for financially supporting L. Junker with a postdoctoral fellowship. This work was also funded by The National Screening Laboratory for the Regional Centers of Excellence in Biodefense and Emerging Infectious Disease (NIH grant U54AI057159) and additional funding from NIH grant CA59021(to J.C.).
Footnotes
Published ahead of print on 30 July 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Banin, E., K. M. Brady, and E. P. Greenberg. 2006. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 72:2064-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochicchio, G. V., M. Joshi, K. Bochicchio, K. Tracy, and T. M. Scalea. 2004. A time-dependent analysis of intensive care unit pneumonia in trauma patients. J. Trauma 36:296-303. [DOI] [PubMed] [Google Scholar]
- 3.Bradley, D. E. 1974. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with non-retractile pili. Virology 58:149-163. [DOI] [PubMed] [Google Scholar]
- 4.Cabezudo, I., R. L. Thompson, R. F. Selden, S. H. Guenthner, and R. P. Wenzel. 1984. Cefsulodin sodium therapy in cystic fibrosis patients. Antimicrob. Agents Chemother. 25:4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatzinikolaou, I., D. Abi-Said, G. P. Body, K. V. Rolston, J. J. Tarrand, and G. Samonis. 2000. Recent experience with Pseudomonas aeruginosa bacteremia in patients with cancer: retrospective analysis of 245 episodes. Arch. Intern. Med. 160:501-509. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 1995:711-745. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 8.Costerton, W., R. Veeh, M. Shirtliff, M. Pasmore, C. Post, and G. Ehrlich. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 112:1466-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crump, J. A., and P. J. Collignon. 2000. Intravascular catheter-associated infections. Eur. J. Clin. Microbiol. Infect. Dis. 19:1-8. [DOI] [PubMed] [Google Scholar]
- 10.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. [DOI] [PubMed] [Google Scholar]
- 11.Dieter, R. S. 2004. Coronary artery stent infection. Catheter Cardiovasc Interv. 62:281. [DOI] [PubMed] [Google Scholar]
- 12.Doggett, R. G. 1969. Incidence of mucoid Pseudomonas aeruginosa from clinical sources. Appl. Environ. Microbiol. 18:936-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doggett, R. G., G. M. Harrison, and E. S. Wallis. 1964. Comparison of some properties of Pseudomonas aeruginosa isolated from infections in persons with and without cystic fibrosis. J. Bacteriol. 87:427-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FitzSimmons, S. C. 1993. The changing epidemiology of cystic fibrosis. J. Pediatr. 122:1-9. [DOI] [PubMed] [Google Scholar]
- 15.Fleiszig, S. M. J., S. K. Arora, R. Van, and R. Ramphal. 2001. FlhA, a component of the flagellum assembly apparatus of Pseudomonas aeruginosa, plays a role in internalization by corneal epithelial cells. Infect. Immun. 69:4931-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Givskov, M., R. de Nys, M. Manefield, L. Gram, R. Maximilien, L. Eberl, S. Molin, P. Steinberg, and S. Kjelleberg. 1996. Eukaryotic interference with homoserine lactone-mediated prokaryotic signaling. J. Bacteriol. 178:6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorman, S. P. 1999. Chance only favours the prepared mind: the future for medical devices? J. Pharm. Pharmacol. 51:49.10197417 [Google Scholar]
- 19.Gram, L., R. de Nys, R. Maximilien, M. Givskov, P. Steinberg, and S. Kjelleberg. 1996. Inhibitory effects of secondary metabolites from the red alga Delisea pulchra on swarming motility of Proteus mirabilis. Appl. Environ. Microbiol. 62:4284-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habash, M., and G. Reid. 1999. Microbial biofilms: their development and significance for medical device-related infections. J. Clin. Pharmacol. 39:887-898. [DOI] [PubMed] [Google Scholar]
- 21.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87-102. [DOI] [PubMed] [Google Scholar]
- 22.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchison, M. L., and J. R. W. Govan. 1999. Pathogenicity of microbes associated with cystic fibrosis. Microbes Infect. 1:1005-1014. [DOI] [PubMed] [Google Scholar]
- 24.Manefield, M., R. de Nys, N. Kumar, R. Read, M. Givskov, P. Steinberg, and S. Kjelleberg. 1999. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145:283-291. [DOI] [PubMed] [Google Scholar]
- 25.Manefield, M., T. B. Rasmussen, M. Henzter, J. B. Andersen, P. Steinberg, S. Kjelleberg, and M. Givskov. 2002. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148:1119-1127. [DOI] [PubMed] [Google Scholar]
- 26.Maschmeyer, G., and I. Braveny. 2000. Review of the incidence and prognosis of Pseudomonas aeruginosa infections in cancer patients in the 1990s. Eur. J. Clin. Microbiol. Infect. Dis. 19:915-925. [DOI] [PubMed] [Google Scholar]
- 27.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 29.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 30.Pretsch, E., P. Bühlmann, and C. Affolter. 2000. Structure determination of organic compounds, Tables of spectral data, 3rd ed. Springer-Verlag, Berlin, Germany.
- 31.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 32.Ramsey, B. W., H. L. Dorkin, J. D. Eisenberg, R. L. Gibson, I. R. Harwood, R. M. Kravitz, D. V. Schidlow, R. W. Wilmott, S. J. Astley, M. A. McBurnie, K. Wentz, and A. L. Smith. 1993. Efficacy of aerosolized tobramycin in patients with cystic fibrosis. N. Engl. J. Med. 328:1740-1746. [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen, T. B., M. Manefield, J. B. Andersen, L. Eberl, U. Anthoni, C. Christophersen, P. Steinberg, S. Kjelleberg, and M. Givskov. 2000. How Delisea pulchra furanones affect quorum sensing and swarming motility in Serratia liquefaciens MG1. Microbiology 146:3237-3244. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, vol. 3, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Shimizu, M., Y. Takase, S. Nakamura, H. Katae, A. N. Minami, K. S. Inoue, M. Ishiyama, and Y. Kubo. 1975. Pipemidic acid, a new antibacterial agent active against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 8:132-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, J.-H., T. D. Y. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.