Abstract
Microcin E492-producing bacteria secrete both unmodified and posttranslationally modified microcins. The modification consists of a C-glucosylated linear trimer of N-(2,3-dihydroxybenzoyl)-l-serine, a catecholate siderophore related to salmochelins and enterobactin. We show here that repression of enterobactin biosynthesis inhibits the acquisition of microcin E492 posttranslational modification, as monitored by high-performance liquid chromatography and mass spectrometry. Furthermore, exogenous enterobactin restored the production of posttranslationally modified microcin in a bacterial strain deficient in enterobactin synthesis. We thus concluded that enterobactin serves as a precursor for the synthesis of the posttranslationally modified microcin and that the unmodified microcin is an incompletely processed form of mature microcin E492. Gene disruption experiments showed that MceC and MceD, two enzymes encoded by the mceABCDEFGHIJ gene cluster, are involved in the synthesis of the microcin E492 posttranslational modification, as followed by mass spectrometry. Genes homologous to iroB and iroD, required for the conversion (linearization and C-glycosylation) of enterobactin into salmochelins, efficiently complemented mceC and mceD, respectively. Based on our results, a model is proposed for the biosynthesis of the mature siderophore-carrying peptide.
Microcin E492 (MccE492) is a pore-forming antimicrobial peptide naturally secreted by Klebsiella pneumoniae RYC492 (14). Its antibacterial activity, which is directed against gram-negative bacteria, with MICs in the range of 0.1 to 1 μM (16), requires the ManYZ inner membrane complex involved in mannose uptake for antibacterial activity (7). The mceABCDEFGHIJ gene cluster responsible for MccE492 production, export, and immunity was cloned from the K. pneumoniae genome into the pJAM229 plasmid (39). Expressed as a recombinant peptide in Escherichia coli VCS257 harboring pJAM229, MccE492 was initially characterized as an unmodified 84-residue peptide (32) deriving from the precursor protein MceA by elimination of a leader peptide. More recently, a modified form of MccE492 endowed with more potent antibacterial activity was isolated from the culture supernatant of the same recombinant E. coli strain as well as from the naturally producing K. pneumoniae strain (33). It was characterized as a siderophore-carrying peptide (siderophore-peptide) resulting from the linkage of a C-glucosylated linear trimer of N-(2,3-dihydroxybenzoyl)-l-serine (DHBS) to the Ser84 carboxylate (33). MccE492 belongs to the recently defined class IIb of microcins (17), encompassing the linear high-molecular-mass microcins that may carry C-terminal siderophores.
Siderophores are molecules designed by bacteria to chelate ferric iron, enabling its uptake into bacteria (for reviews, see references 2 and 37). In gram-negative bacteria, ferric siderophore complexes are recognized by specific outer membrane receptors and then transferred into the cytoplasm by periplasmic binding proteins and inner membrane transporters. Among these chelating agents, the recently characterized salmochelin S4 derives from enterobactin, a cyclic trimer of DHBS, by the occurrence of two β-d-glucose (Glc) moieties linked to the DHBS units through C-glycosidic bonds (8). The production of salmochelins by Salmonella enterica and uropathogenic E. coli occurs under iron-poor conditions and is dependent on enterobactin synthesis and the iroBCDEN gene cluster (27). While iroC and -N are involved in siderophore transport (27, 41), the three remaining genes, iroB, -D, and -E, are involved in the conversion of enterobactin into salmochelin S4. IroB is an enterobactin C-glucosyltransferase able to catalyze the transfer of glucose from UDP-glucose to enterobactin in vitro (21). IroD and IroE are ferric enterobactin esterase-like proteins (41). While both are able to convert salmochelin S4 into the linear trimer salmochelin S2, IroD degrades further salmochelin S2 into salmochelins S1, SX, and DHBS (30, 41).
Synthesis of the MccE492 posttranslational modification was shown to be dependent on the culture medium composition. Indeed, the addition of Casamino Acids to M63 medium led to the major production of the unmodified peptide, whereas the addition of a trypsin digest of casein favored the production of the siderophore-peptide (33). Because the MccE492 posttranslational modification is reminiscent of salmochelins and because salmochelin synthesis is dependent on enterobactin, we predicted that factors impairing enterobactin synthesis might negatively control the synthesis of the microcin as a siderophore-peptide. We therefore studied the culture parameters, including growth phase and medium composition, which might prevent microcin posttranslational processing. We demonstrated that inhibition of enterobactin synthesis by a high iron concentration or free aromatic amino acids correlated with a breakdown of MccE492 posttranslational modification. In addition, exogenous enterobactin restored the microcin posttranslational modification in a strain deficient in enterobactin synthesis. Since enterobactin was required for MccE492 posttranslational modification, we examined the role of mceC and mceD, which are iroB and iroD homologues (15), respectively, in this process. This was performed through gene disruption and functional complementation. From our data, we concluded that the siderophore-carrying microcin is the actual mature peptide. Consequently, we termed MccE492 the mature microcin (the siderophore-peptide previously referred to as MccE492m [33]), while the unmodified microcin lacking the N-terminal leader (initially referred to as MccE492 [32]) was renamed unmodified MccE492 (u-MccE492). Based on our results, we propose a model for the biosynthesis of the MccE492 posttranslational modification.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used for this study are described in Table 1. Microcin production was performed with E. coli MC4100, E. coli RYC1000 (a generous gift from F. Moreno), E. coli VCS257, or E. coli C600 aroB (a generous gift from P. Boulanger). The plasmid used for standard production of MccE492 was pJAM229 (39). E. coli strains were freshly transformed with pJAM229, pGV100, or pGV200 (this study). For complementation studies, pGV100 and/or pGV200 was cotransfected with pGV300 (this study) or pEX100 (a generous gift from M. Laviña). Transformants were grown in M63 minimal medium containing 0.25% glucose, 0.25 g/liter MgSO4, and 1 mg/liter thiamine. Depending on the experiment, M63 medium was supplemented with either 1 g/liter Casamino Acids (Difco), 1 g/liter tryptone (Fluka, Biochemika), 1 mM l-tyrosine, 1 mM l-tryptophan, 1 mM l-phenylalanine, or a mixture of Phe, Trp, and Tyr (1 mM [each]). All amino acids were purchased from Sigma (biotechnology grade). In other experiments, enterobactin (2.5 μM) (EMC Microcollections GmbH, Tübingen, Germany) or FeCl3 (16 or 200 μM soluble iron concentration) was added. Antibiotics were used at the following concentrations: ampicillin and kanamycin, 50 mg/liter; and chloramphenicol, 30 mg/liter. Cultures were routinely performed for 15 h at 37°C with vigorous shaking (250 rpm).
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Relevant features | Reference or source |
|---|---|---|
| E. coli strains | ||
| C600 aroB | F−thr leu fhuA lacY thi supE aroB | Laboratory collection |
| MC4100 | araD139 Δ(argF lac)U169 rpsL relA flbB deoC | Laboratory collection |
| VCS257 | ton-53 dapD8 lacY1 glnV44 | Stratagene |
| RYC1000 | MC4100 Δrbs-7 recA56 gyrA | 23 |
| Plasmids | ||
| pET19b | His tag fusion vector; Ampr | Novagen |
| pET28b | His tag fusion vector; Kanr | Novagen |
| pEX100 | pACYC184 derivative carrying MccH47 gene cluster; Chlr | 22 |
| pGV100 | pJAM229 mceD; Ampr | This study |
| pGV200 | pJAM229 mceC; Ampr | This study |
| pGV300 | pET28b derivative carrying iroD; Kanr | This study |
| pJAM229 | Plasmid carrying MccE492 gene cluster; Ampr | 39 |
| pMZ2038 | pET19b derivative carrying iroD; Ampr | 41 |
Gene disruption and heterologous complementation.
pJAM229 was digested with SpeI at one single site within the mceD gene. A stop codon was inserted at the restriction site by cloning an oligonucleotide duplex complementary to the SpeI-generated cohesive ends into the open vector. The duplex was generated with two complementary 5′-phosphorylated oligonucleotides, 5′-CTAGGGTAATCTAGACTATTACC-3′ and 5′-CTAGGGTAATAGTCTAGATTACC-3′, designed to abolish the SpeI site after insertion. Prior to transformation, the ligation product was digested with SpeI in order to open pJAM229 vectors lacking the insert. The positive clones were then selected by simple restriction map comparison, with all positive clones being uncleaved upon SpeI digestion. One positive clone containing an inactivated mceD gene was named pGV100 and selected for microcin expression studies. A similar strategy was used for mceC disruption. Basically, pJAM229 was digested with RsrII and ligated to a stop codon-containing duplex generated with the 5′-phosphorylated oligonucleotides 5′-GTCTAAGCGGCCGCTTATA-3′ and 5′-GTCTATAAGCGGCCGCTTA-3′. Prior to transformation, the ligation product was digested with RsrII and the positive clones were selected by restriction map comparison. One positive clone containing an inactivated mceC gene was selected and named pGV200. For mceD complementation, bacteria were transformed with the iroD-carrying pGV300 plasmid. This plasmid was obtained by subcloning the E. coli iroD gene from pMZ2038 (a generous gift from M. Zhu and K. Hantke) (41) into pET28b. Subcloning was achieved upon pMZ2038 double digestion with NdeI and XhoI and ligation into the same sites of the pET28b polylinker. Complementation studies were also performed with pEX100, which carries the MccH47 gene cluster (22).
Microcin purification.
Culture supernatants were separated from bacterial cells by centrifugation (6,000 × g, 15 min, 4°C) and subjected to solid-phase extraction on a Sep-Pak C8 cartridge (Waters Corp.) preequilibrated with 0.1% aqueous trifluoroacetic acid (TFA). Cartridges were washed with 0.1% aqueous TFA prior to successive elution with 30%, 35%, and 40% acetonitrile in 0.1% aqueous TFA. Detection of MccE492-specific antibacterial activity in Sep-Pak fractions was performed by radial diffusion assays, as described below. The 40% Sep-Pak fraction containing microcins was lyophilized and analyzed by reversed-phase high-performance liquid chromatography (RP-HPLC) on an Inertsil ODS2 column (5 μm, 4.6 mm by 250 mm; Interchim, France). Separation was performed at a flow rate of 1 ml/min under isocratic conditions with 40% acetonitrile in 0.1% aqueous TFA. Absorbance was monitored at 226 nm, and fractions were hand collected.
Antibacterial assays.
A gel overlay was prepared by inoculating 12 ml M63 medium (containing 6.5 g/liter agar) with 107 CFU/ml of bacteria (either E. coli MC4100 or E. coli MC4100/pJAM229) during the exponential phase of growth. Petri dishes containing 20 ml M63 solid medium (15 g/liter agar) were overlaid with the bacterial suspension. After solidification, 10 μl of the fraction to be analyzed was placed on the overlay. After a 16-h incubation at 37°C, plates were analyzed for the presence of inhibition halos. Fractions inhibitory to E. coli MC4100 but not to E. coli MC4100/pJAM229 were considered to contain MccE492/u-MccE492.
MALDI-TOF-MS.
Microcin-containing fractions were analyzed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF-MS) as previously described (9), using a Voyager-De-Pro MALDI-TOF mass spectrometer (Applied Biosystems), with α-cyano-4-hydroxycinnamic acid as the matrix.
Soluble iron quantification.
A colorimetric assay was performed to quantify soluble iron in the culture medium. The total iron concentration was determined after reduction of Fe(III) to Fe(II). Reduction was performed by diluting the culture medium 1:3 (vol/vol) in 1.65 M sodium acetate in 50% acetic acid and adding 18 g/liter hydroxylamine chlorhydrate. Complex formation of Fe(II) was then obtained by adding 0.9 g/liter orthophenanthroline. The absorbance at 510 nm was measured with a Uvikon 932 spectrophotometer (Kontron Instruments) after a 15-min incubation at room temperature and compared to that on a standard curve obtained with Fe2(SO4)3, (NH4)2SO4, 24 H2O.
RESULTS
Synthesis of MccE492 posttranslational modification is not stationary phase dependent.
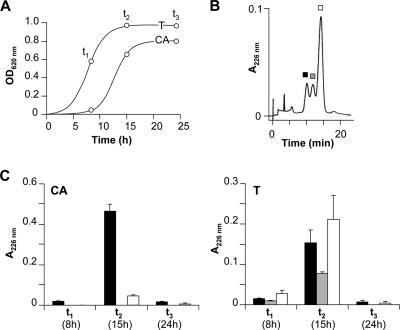
We have previously shown that growth of E. coli VCS257 harboring pJAM229 is delayed when Casamino Acids are used instead of tryptone to supplement M63 medium (33). In order to determine whether posttranslational modification of MccE492, which is mainly observed in tryptone-containing M63 medium (M63-T medium) (33), is stationary phase dependent, we performed time course experiments with both culture media. Aliquot fractions were removed at different bacterial phases of growth, i.e., 8 h (t1), 15 h (t2), and 24 h (t3) (Fig. 1A). In three independent experiments, the culture supernatants were fractionated on a reversed-phase Sep-Pak cartridge and analyzed for the presence of MccE492 versus u-MccE492 (Fig. 1B). RP-HPLC and MALDI-TOF-MS analysis of the 40% Sep-Pak fractions revealed the presence of both u-MccE492 (lowest retention time; 7,887 Da) and MccE492 (highest retention time; 8,718 Da) in most of the fractions analyzed (Fig. 1B and C and 2). For M63-T culture medium, chromatograms for t1 (Fig. 1B) and t2 displayed an additional peak (intermediary retention time). This peak corresponded to species at m/z 8,273 and 8,496, previously identified as intermediate forms of MccE492 carrying one and two DHBS units, respectively (33) (Fig. 2). In both culture media, the overall microcin production was maximal at t2 and dropped dramatically at t3 (Fig. 1C). However, different microcin production patterns were observed under both culture conditions. In Casamino Acid-containing medium (M63-CA medium), minor amounts of the MccE492 siderophore-peptide were found independent of the growth phase. In M63-T medium, the production of MccE492 was similar (not statistically different) to that of u-MccE492 all over the time course (Fig. 1C). It was therefore concluded from both experiments that the biosynthesis of the MccE492 posttranslational modification is not stationary phase dependent.
FIG. 1.
Pattern of microcin production over the growth phases. (A) Cultures of E. coli VCS257/pJAM229 in M63 medium containing either Casamino Acids (CA) or tryptone (T) were analyzed for microcin production (open circles) at different steps of bacterial growth (t1, t2, and t3), monitored by optical density measurements at 620 nm (OD620). (B) Typical RP-HPLC profile of the 40% Sep-Pak fraction resulting from the M63-T culture supernatant at t2. Separation was performed on an ODS2 Inertsil column under 40% acetonitrile in 0.1% aqueous TFA. MS experiments on the three fractions revealed the following molecular masses: 7,887 Da (black square), 8,272 and 8,495 Da (gray square), and 8,718 Da (white square). (C) The average absorbance for every peak is presented for t1, t2, and t3. Microcin species are represented with the same code as that for panel B. Values are presented as the means for three independent experiments, with standard errors of the means.
FIG. 2.
Structure of MccE492 and derivatives. MccE492 (8,718 Da) is a posttranslationally modified 84-residue peptide. The modification consists of a trimer of DHBS linked via a C-glycosidic bond to a β-d-glucose moiety that is itself linked to Ser84 through an O-glycosidic bond. u-MccE492 (7,887 Da) is the unmodified 84-residue peptide. u-MccE492-Glc-DHBS (8,272 Da) and u-MccE492-Glc-DHBS2 (8,495 Da) correspond to intermediate forms carrying a β-d-glucose moiety linked to one DHBS and two DHBS moieties, respectively. The amino acid sequence is indicated in italics. The mass differences between the different structures are shown at the top.
High iron concentration inhibits biosynthesis of the MccE492 posttranslational modification.
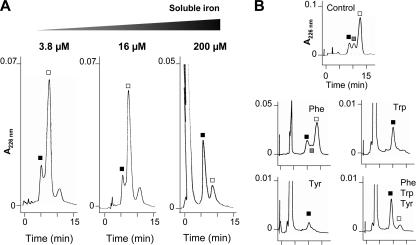
The total iron concentrations in microcin culture media were measured as 9.10 ± 2.53 μM in M63-CA medium and 3.77 ± 0.04 μM in M63-T medium. In order to determine whether the greater synthesis of MccE492 in M63-T medium was a consequence of a lower iron concentration, FeCl3 was added to the M63-T medium in order to reach a 16 μM soluble iron concentration, i.e., higher than the concentration found in M63-CA medium. The 40% Sep-Pak fractions issued from the 15-h culture supernatants were subjected to RP-HPLC and MALDI-TOF-MS. The chromatograms obtained were identical for regular (3.8 μM) and iron-loaded (16 μM) cultures (Fig. 3A), with a major absorbance peak corresponding to MccE492 (8,718 Da) preceded by a minor absorbance peak corresponding to u-MccE492 (7,887 Da). However, when the soluble iron concentration was increased up to 200 μM in M63-T medium (Fig. 3A), the less-retained absorbance peak became largely predominant, indicating that under these conditions the microcin was secreted mainly as an unmodified peptide.
FIG. 3.
Inhibition of biosynthesis of the MccE492 posttranslational modification by high iron concentrations and free aromatic amino acids. RP-HPLC analysis was performed with the 40% Sep-Pak fractions issued from culture supernatants. Separation was performed under 40% acetonitrile in 0.1% aqueous TFA. The molecular masses detected in the fractions were 7,887 Da (black squares), 8,272 and 8,495 Da (gray squares), and 8,718 Da (white squares). (A) Iron. E. coli VCS257/pJAM229 was cultured in M63-T medium (3.8 μM soluble iron) or in the same medium supplemented with FeCl3 to reach 16 or 200 μM iron. (B) Aromatic amino acids. E. coli MC4100/pJAM229 was cultured in M63 medium without amino acids (control) or supplemented with either 1 mM Phe, 1 mM Trp, 1 mM Tyr, or a mixture of the three amino acids (Phe, Trp, and Tyr at 1 mM [each]).
Free aromatic amino acids inhibit biosynthesis of the MccE492 posttranslational modification.
One of the major differences between tryptone and Casamino Acids is the process of casein hydrolysis. In the former case, trypsin generates peptides, whereas in the latter case, acid hydrolysis generates free amino acids, which can inhibit several biosynthetic pathways, including that of enterobactin (36). Particular attention was paid to the aromatic amino acids, which share with enterobactin the same initial steps of biosynthesis in the shikimate pathway (6). Therefore, the microcin-producing strain was cultured in M63 medium without added amino acids. The various forms of microcin secreted were compared to those obtained in M63 medium supplemented with Phe, Trp, Tyr, or a mixture of the three amino acids. Consistent with earlier reports (4, 5), the addition of Trp or Tyr to the culture medium had a negative effect on bacterial growth, which was less pronounced when Phe was added. In the absence of free amino acids, the 40% Sep-Pak fraction issued from the M63 culture supernatant displayed an RP-HPLC chromatogram (Fig. 3B, control panel) similar to that obtained with M63-T medium (Fig. 1B). MALDI-TOF-MS analyses confirmed that the three peaks corresponded to u-MccE492, the intermediate forms described above, and MccE492 (Fig. 3B, control panel). Whereas the addition of Phe did not significantly modify the pattern of microcins secreted, with MccE492 being largely predominant, Trp and Tyr dramatically shifted the production towards u-MccE492, with one single peak detected at the lowest retention time (Fig. 3B). Similarly, the combined addition of Phe, Trp, and Tyr led to the major production of u-MccE492, with only residual amounts of MccE492 (Fig. 3B). Altogether, these results indicate that free Trp and/or Tyr added to M63 culture medium inhibits the synthesis of the MccE492 posttranslational modification.
Exogenous enterobactin restores biosynthesis of the MccE492 posttranslational modification in an enterobactin-deficient E. coli strain.
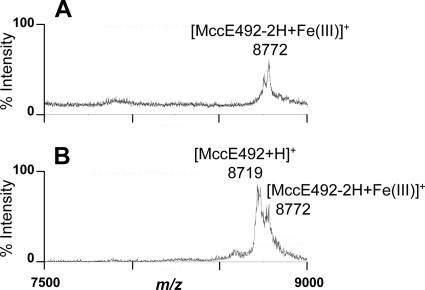
To definitely demonstrate whether enterobactin synthesis is the limiting factor for MccE492 biosynthesis, E. coli C600 aroB harboring pJAM229, a strain deficient in the production of both enterobactin and the MccE492 siderophore-peptide (33), was cultured in the presence of 2.5 μM exogenous enterobactin. The 40% Sep-Pak fractions issued from the culture of E. coli C600 aroB pJAM229 in M63-T medium loaded with exogenous enterobactin were analyzed by MALDI-TOF-MS. These fractions almost lacked the ion corresponding to u-MccE492, but instead displayed one major ion at m/z 8,772 (increment of 54 Da from MccE492) (Fig. 4A). This was in contrast to the case with cultures of the same E. coli C600 aroB/pJAM229 strain in M63-T medium lacking exogenous enterobactin, which contained one single species corresponding to u-MccE492 (MH+ at m/z 7,888) (33). Interestingly, the MALDI-TOF-MS spectrum of MccE492 mixed with FeCl3 showed an ion at m/z 8,772 (Fig. 4B) characteristic of the cationized species [MccE492-2H+Fe(III)]+. This indicates that the m/z 8,772 ion detected for the enterobactin-loaded cultures most likely corresponds to MccE492 (MH+ at m/z 8,718) cationized with Fe(III). Therefore, exogenous ferric enterobactin appears to restore the synthesis of the MccE492 siderophore-peptide in enterobactin-deficient E. coli C600 aroB/pJAM229.
FIG. 4.
Restoration of biosynthesis of the MccE492 posttranslational modification in an E. coli strain deficient in enterobactin synthesis, as shown by MALDI-TOF-MS. (A) Analysis of the 40% Sep-Pak fraction issued from the culture supernatant of E. coli C600 aroB cultured in M63-T medium in the presence of 2.5 μM enterobactin. The broad peak below m/z 8,000 corresponds to u-MccE492. (B) Analysis of purified MccE492 incubated with FeCl3 (FeCl3/MccE492 ratio, 4:1).
MceC and MceD are required for biosynthesis of the MccE492 posttranslational modification.
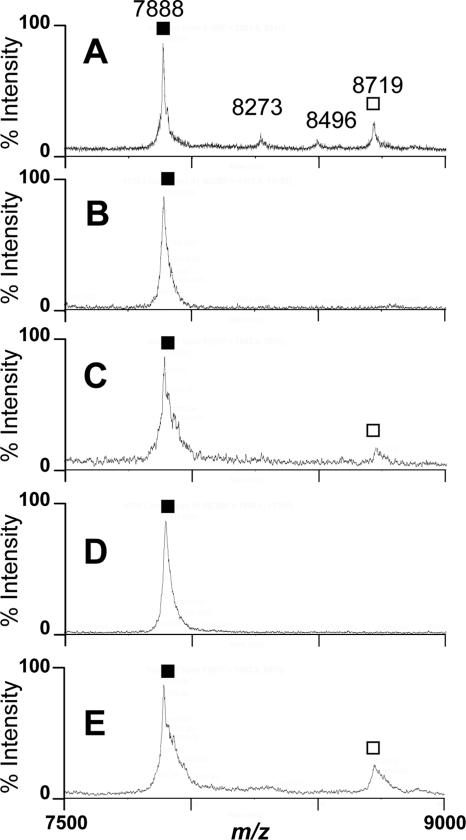
MceC and MceD show major sequence homology with E. coli IroB (75% identity) and IroD (57% identity) (15), a C-glycosyltransferase and a ferric enterobactin esterase involved in salmochelin biosynthesis, respectively (8, 30, 41). In order to examine their potential role in the biosynthesis of the MccE492 siderophore-peptide, we disrupted their respective genes by inserting a stop codon-containing oligonucleotide duplex at chosen sites in the pJAM229 plasmid. The resulting plasmids were then used to transform E. coli RYC1000. In order to favor the production of modified microcin, the strains were cultured under nonrepressive conditions. The microcin species were extracted, as described above, from culture supernatants of E. coli RYC1000 harboring pGV100 (pJAM229 mceD), pGV200 (pJAM229 mceC), or pJAM229 (control). MALDI-TOF-MS analyses of the 40% Sep-Pak fractions revealed that both mceC and mceD mutant strains secreted u-MccE492 only (Fig. 5B and D), but in very small amounts, as monitored by RP-HPLC (data not shown). In contrast, MccE492 and intermediate forms with various degrees of modification were found in the control supernatants (Fig. 5A). This indicates that both genes are required for MccE492 posttranslational modification. Actually, it was observed that both mceC and mceD gene disruption did not affect E. coli growth but resulted in a dramatic drop in the microcin concentration in culture supernatants.
FIG. 5.
Inactivation and restoration of biosynthesis of the MccE492 posttranslational modification upon mceD or mceC gene disruption and complementation with pEX100. MALDI-TOF-MS spectra are shown for the 40% Sep-Pak fractions issued from culture supernatants of E. coli RYC1000 harboring pJAM229 (A; control), pGV200 (B; pJAM229 mceC), or pGV100 (D; pJAM229 mceD). In complementation assays, E. coli RYC1000 harbored the MccH47-encoding plasmid pEX100 together with pGV200 (C) or pGV100 (E). White and black symbols indicate the MH+ ions relative to MccE492 and u-MccE492, respectively.
mchA and mchS1 complement mceC and mceD.
In a first attempt, E. coli RYC1000 was cotransfected with pGV100 and the IroD-encoding pGV300 plasmid. However, clones carrying both plasmids were unable to grow in liquid broth, hampering demonstration of the MceD function. Consequently, we assessed whether genes homologous to iroB and iroD required for MccE492 posttranslational modification could be complemented by similar genes found in gene clusters encoding class IIb microcins (for a review, see reference 17). Plasmids carrying disrupted mceC and mceD were consequently transfected into E. coli RYC1000 harboring pEX100, the MccH47-encoding plasmid that carries mchA and mchS1, two genes similar to iroB and iroD, respectively (3). In both cases, mature MccE492 was detected in the 40% Sep-Pak fractions of culture supernatants (Fig. 5C and E), which displayed MALDI-TOF spectra similar to that of the control (Fig. 5A). Therefore, mchA and mchS1 complement mceC and mceD, respectively, which indicates that they encode proteins with similar functions.
DISCUSSION
The biosynthesis of MccE492 has been a puzzling question since the first description of its gene cluster (39). We demonstrated here for the first time that the posttranslational modification of MccE492 (i.e., the addition of a C-glycosylated trimer of DHBS at the C-terminal end of the peptide backbone) requires enterobactin, which is used as a precursor by MceC and MceD, a glycosyltransferase and an enterobactin esterase, respectively, encoded by the MccE492 gene cluster.
This article shows that the synthesis of the MccE492 posttranslational modification is inhibited by conditions that also inhibit enterobactin synthesis. Indeed, free aromatic amino acids hamper the production of MccE492 as a siderophore-peptide. Interestingly, free aromatic amino acids control the shikimate pathway, which leads to the biosynthesis of both enterobactin and aromatic amino acids, at two different steps (Fig. 6). First, Tyr, Phe, and Trp repress the expression of aroF, -G, and -H, respectively (12, 24, 25), which are involved in the first step of the shikimate pathway (35). Second, Trp and Tyr are involved in the synergistic repression of aroL (19, 28), which encodes, together with aroK (38), the shikimate kinase. It can therefore be inferred that the inhibitory effect of a mixture of Phe, Trp, and Tyr on the synthesis of the MccE492 posttranslational modification results from inhibition of the shikimate pathway. Because inhibition of the synthesis of the MccE492 posttranslational modification was observed with Trp and Tyr, but not with Phe alone, aroL was supposed to be the affected gene (Fig. 6). A direct inference from this result is that the lack of posttranslationally modified MccE492 in M63-CA medium is most likely due to control of the shikimate pathway by free Tyr, because Trp is destroyed during the casein hydrolysis process.
FIG. 6.
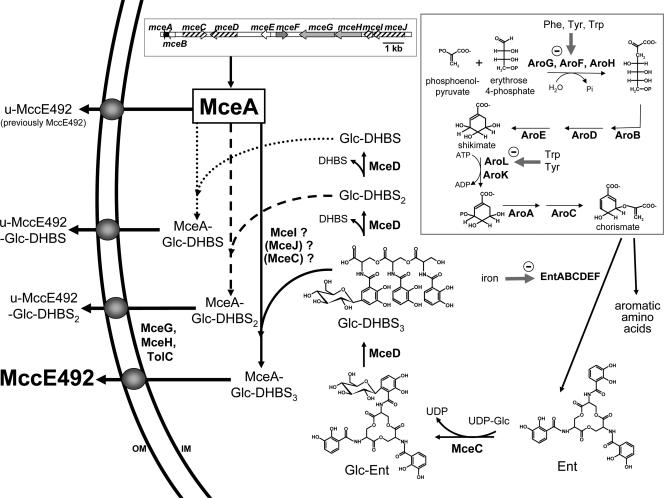
Model for biosynthesis of MccE492 showing that enterobactin is a precursor and that the MccE492 siderophore-peptide is the mature form of the microcin. After translation, MceA (the MccE492 precursor protein) is subjected to posttranslational modification at the C-terminal serine. The synthesis of enterobactin (Ent) is performed by the aroABCDEFGHKL (shikimate pathway; boxed)- and entABCDEF-encoded machineries. Ent is the precursor of posttranslational modification biosynthesis, which most likely starts with MceC, a putative C-glucosyltransferase. The resulting Glc-Ent is then converted into linear Glc-DHBS3 by MceD, a putative enterobactin esterase, and transferred onto the carboxylate of the MceA C-terminal serine. This last step is believed to involve MceI, MceJ, and maybe MceC. MccE492 then results from the cleavage of the N-terminal leader peptide of MceA-Glc-DHBS3 during export (the MceG/MceH/TolC export machinery is represented by gray spheres in the membrane). MccE492 intermediate forms (u-MccE492-Glc-DHBS and u-MccE492-Glc-DHBS2) are thought to result from further hydrolysis of MceA-Glc-DHBS3 (MceD enzymatic activity), cleavage of the leader peptide, and subsequent export. u-MccE492 results from MceA leader cleavage and subsequent export under certain culture conditions. The mceABCDEHGHIJ gene cluster is presented (inset). Genes involved (or supposed to be) in the synthesis, posttranslational modification, and export of MccE492 are shown as plain black arrows, black hatched arrows, and gray arrows, respectively. IM and OM, inner membrane and outer membrane, respectively.
Besides free aromatic amino acids, a high iron concentration (200 μM) was shown to inhibit the synthesis of the MccE492 posttranslational modification. Iron-regulated gene expression in E. coli is largely mediated by Fur, a ferrous iron-binding protein that binds to the so-called Fur box (5′-GATAATGATAATCATTATC-3′) (20) and blocks iron-regulated promoters in a metal-dependent fashion (26). Such iron-regulatory regions are found upstream from mceC and -D within the MccE492 gene cluster (29) as well as upstream from the entCEBA gene cluster (10) required for the conversion of chorismate into enterobactin (Fig. 6). Thus, MccE492 production is most likely controlled by iron through the repression of genes encoding both MccE492 modification enzymes and enterobactin synthesis. The addition of iron to M63-T medium in order to reach a final concentration beyond that in M63-CA medium did not alter the MccE492-to-u-MccE492 ratio, which indicates that the higher iron concentration in M63-CA medium was not responsible for the observed inhibition of synthesis of the MccE492 posttranslational modification. Therefore, the inhibition of MccE492 maturation is most likely mediated by free aromatic amino acids, although it cannot be ruled out that other factors contained in Casamino Acids might be involved (see above). Altogether, our data show that u-MccE492 is an incompletely processed form of the mature MccE492 siderophore-peptide and that its expression is favored under certain culture conditions.
One major finding from this article is that enterobactin is used as a precursor for the biosynthesis of the siderophore-peptide. Indeed, exogenous enterobactin is able to restore the synthesis of the MccE492 posttranslational modification in a strain deficient in the synthesis of both enterobactin and mature MccE492. Therefore, upon inhibition and/or abolition of enterobactin synthesis, E. coli strains harboring the pJAM229 plasmid are able to import ferric enterobactin and to use it as a precursor for MccE492 posttranslational modification. The latter process was shown here to require mceC and mceD from the MccE492 gene cluster. Indeed, inactivation of either mceC or mceD, encoding an enzyme homologous to IroB or IroD involved in salmochelin biosynthesis (8, 30, 41), is sufficient to completely inhibit MccE492 maturation. A posttranslational modification identical to that obtained with the wild-type gene cluster can be restored upon heterologous complementation of the mceC or mceD disrupted gene with the MccH47 gene cluster. It is therefore very likely that the occurrence of genes homologous to iroB and iroD in gene clusters encoding MccE492, MccH47, MccM, and MccI47 (17, 31) enables the synthesis of a siderophore-type posttranslational modification. Based on our findings, a sequence of events can be proposed for the biosynthesis of the MccE492 posttranslational modification (Fig. 6), as follows: (i) as a precursor for MccE492 modification, enterobactin (or exogenous ferric enterobactin) would be C-glucosylated by MceC in an IroB-like manner (21); (ii) the C-glucosylated enterobactin (Glc-Ent) would be hydrolyzed by MceD, a homologue of IroD; (iii) the resulting glucosylated linear trimer of DHBS (Glc-DHBS3) would then be transferred onto MceA (the MccE492 precursor) by enzymes encoded by the MccE492 gene cluster (MceI and, putatively, MceC and MceJ); and (iv) finally, MceA-Glc-DHBS3 would be converted into MccE492 through removal of the leader peptide concomitant with microcin secretion.
Since intact enterobactin, rather than its linear forms, is believed to be the relevant substrate for IroB in vivo (21), we suggest that the MceC-dependent C-glucosylation of enterobactin precedes the MceD-mediated hydrolysis step. In addition, because MceI shares homology with acyltransferases involved in the activation of RTX toxins from gram-negative bacteria (11, 34), it could catalyze the acylation of β-d-glucose by the C-terminal serine of MceA. This hydrolysis step is likely to also involve MceJ, whose gene is cotranscribed with mceI (29) and is required for the detectable production/secretion of MccE492 (13). However, it cannot be ruled out that this step also requires MceC, which like its UrdGT2 homologue from Streptomyces fradiae (18), may display both C- and O-glycosyltransferase activities. Since cleavage of the leader peptide of class II microcins is concomitant with export (17), MceA is likely converted into MceA-Glc-DHBS3 prior to the cleavage of the leader peptide and microcin secretion. Processing of MceA-Glc-DHBS3 most probably requires the N-terminal proteolytic domain of MceG, which is involved, together with MceH and the chromosome-encoded TolC protein, in the export of MccE492 (for a review, see reference 17). Because u-MccE492 as well as intermediate forms that carry only one or two DHBS units (Fig. 2) is found in the culture medium, the MceG/MceH/TolC export machinery is proposed to recognize not only MceA-Glc-DHBS3 but also MceA, MceA-Glc-DHBS, and MceA-Glc-DHBS2. The intermediate forms are proposed to result from the transfer of Glc-DHBS and Glc-DHBS2, which would be generated by an MceD-mediated hydrolysis of Glc-DHBS3 (Fig. 6), onto MceA, similar to salmochelin hydrolysis by IroD (30). However, it cannot be ruled out that they result from the MceD-mediated hydrolysis of MceA-Glc-DHBS3.
In summary, MceA and enterobactin have been shown here to be the substrates of the MccE492 enzyme machinery for the biosynthesis of the MccE492 siderophore-peptide. Accordingly, u-MccE492 is an incompletely processed microcin that is secreted and accumulates in the culture medium under specific conditions. Evidence has shown that (i) the gene clusters of class IIb microcins that carry genes homologous to mceC, mceD, mceI, and mceJ are able to complement each other (31; this study); and (ii) enterobactin is required for their biosynthesis (3, 33; this study). We therefore propose that the model presented here for MccE492 biosynthesis more generally applies to class IIb microcins, provided that the producing strain is proficient in enterobactin synthesis and that the entire microcin-dedicated enzyme machinery is encoded by the microcin gene cluster.
Acknowledgments
We thank Sophie Duquesne for her support in molecular biology and Gérard Gastine for his precious technical assistance in microbiology. We also thank Mingang Zhu and Klaus Hantke (Universität Tübingen, Germany) and Magela Laviña (Facultad de Ciencias, Montevideo, Uruguay) for the generous gifts of pMZ2038 and pEX100, respectively. We are grateful to Felipe Moreno (Hospital Ramòn y Cajal, Madrid, Spain) for E. coli strain RYC1000 and to Pascale Boulanger (CNRS-UMR 8619, Orsay, France) for the E. coli C600 aroB strain. We thank the mass spectrometry facility (Plate-Forme de Spectrométrie de Masse) at the National Museum of Natural History.
Footnotes
Published ahead of print on 23 July 2007.
REFERENCES
- 1.Reference deleted.
- 2.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 3.Azpiroz, M. F., and M. Laviña. 2004. Involvement of enterobactin synthesis pathway in production of microcin H47. Antimicrob. Agents Chemother. 48:1235-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beerstecher, E. J., and W. Shive. 1947. Tryptophan as a competitive growth inhibiting analog of phenylalanine. J. Am. Chem. Soc. 69:461-462. [DOI] [PubMed] [Google Scholar]
- 5.Beerstecher, E. J., and W. Shive. 1947. Prevention of phenylalanine synthesis by tyrosin. J. Biol. Chem. 167:527-534. [PubMed] [Google Scholar]
- 6.Bentley, R. 1990. The shikimate pathway—a metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25:307-384. [DOI] [PubMed] [Google Scholar]
- 7.Bieler, S., F. Silva, C. Soto, and D. Belin. 2006. Bactericidal activity of both secreted and nonsecreted microcin E492 requires the mannose permease. J. Bacteriol. 188:7049-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bister, B., D. Bischoff, G. J. Nicholson, M. Valdebenito, K. Schneider, G. Winkelmann, K. Hantke, and R. D. Sussmuth. 2004. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals 17:471-481. [DOI] [PubMed] [Google Scholar]
- 9.Blond, A., M. Cheminant, D. Destoumieux-Garzón, I. Ségalas-Milazzo, J. Peduzzi, C. Goulard, and S. Rebuffat. 2002. Thermolysin-linearized microcin J25 retains the structured core of the native macrocyclic peptide and displays antimicrobial activity. Eur. J. Biochem. 269:6212-6222. [DOI] [PubMed] [Google Scholar]
- 10.Brickman, T. J., B. A. Ozenberger, and M. A. McIntosh. 1990. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J. Mol. Biol. 212:669-682. [DOI] [PubMed] [Google Scholar]
- 11.Burrows, L. L., and R. Y. Lo. 1992. Molecular characterization of an RTX toxin determinant from Actinobacillus suis. Infect. Immun. 60:2166-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cobbett, C. S., and M. L. Delbridge. 1987. Regulatory mutants of the aroF-tyrA operon of Escherichia coli K-12. J. Bacteriol. 169:2500-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corsini, G., M. Baeza, O. Monasterio, and R. Lagos. 2002. The expression of genes involved in microcin maturation regulates the production of active microcin E492. Biochimie 84:539-544. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo, V., and A. P. Pugsley. 1985. Microcin E492, a low-molecular-weight peptide antibiotic which causes depolarization of the Escherichia coli cytoplasmic membrane. Antimicrob. Agents Chemother. 27:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Destoumieux-Garzón, D., J. Péduzzi, X. Thomas, C. Djediat, and S. Rebuffat. 2006. Parasitism of iron-siderophore receptors of Escherichia coli by the siderophore-peptide microcin E492m and its unmodified counterpart. Biometals 19:181-191. [DOI] [PubMed] [Google Scholar]
- 16.Destoumieux-Garzón, D., X. Thomas, M. Santamaria, C. Goulard, M. Barthelemy, B. Boscher, Y. Bessin, G. Molle, A. M. Pons, L. Letellier, J. Peduzzi, and S. Rebuffat. 2003. Microcin E492 antibacterial activity: evidence for a TonB-dependent inner membrane permeabilization on Escherichia coli. Mol. Microbiol. 49:1031-1041. [DOI] [PubMed] [Google Scholar]
- 17.Duquesne, S., D. Destoumieux-Garzón, J. Peduzzi, and S. Rebuffat. 2007. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat. Prod. Rep. 24:708-734. [DOI] [PubMed] [Google Scholar]
- 18.Durr, C., D. Hoffmeister, S. E. Wohlert, K. Ichinose, M. Weber, U. Von Mulert, J. S. Thorson, and A. Bechthold. 2004. The glycosyltransferase UrdGT2 catalyzes both C- and O-glycosidic sugar transfers. Angew. Chem. Int. Ed. Engl. 43:2962-2965. [DOI] [PubMed] [Google Scholar]
- 19.Ely, B., and J. Pittard. 1979. Aromatic amino acid biosynthesis: regulation of shikimate kinase in Escherichia coli K-12. J. Bacteriol. 138:933-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischbach, M. A., H. Lin, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proc. Natl. Acad. Sci. USA 102:571-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaggero, C., F. Moreno, and M. Lavina. 1993. Genetic analysis of microcin H47 antibiotic system. J. Bacteriol. 175:5420-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genilloud, O., M. C. Garrido, and F. Moreno. 1984. The transposon Tn5 carries a bleomycin-resistance determinant. Gene 32:225-233. [DOI] [PubMed] [Google Scholar]
- 24.Gibson, F., and J. Pittard. 1968. Pathways of biosynthesis of aromatic amino acids and vitamins and their control in microorganisms. Bacteriol. Rev. 32:465-492. [PMC free article] [PubMed] [Google Scholar]
- 25.Grove, C. L., and R. P. Gunsalus. 1987. Regulation of the aroH operon of Escherichia coli by the tryptophan repressor. J. Bacteriol. 169:2158-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 27.Hantke, K., G. Nicholson, W. Rabsch, and G. Winkelmann. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 100:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heatwole, V. M., and R. L. Somerville. 1992. Synergism between the Trp repressor and Tyr repressor in repression of the aroL promoter of Escherichia coli K-12. J. Bacteriol. 174:331-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagos, R., M. Baeza, G. Corsini, C. Hetz, E. Strahsburger, J. A. Castillo, C. Vergara, and O. Monasterio. 2001. Structure, organization and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol. Microbiol. 42:229-243. [DOI] [PubMed] [Google Scholar]
- 30.Lin, H., M. A. Fischbach, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 127:11075-11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poey, M. E., M. F. Azpiroz, and M. Laviña. 2006. Comparative analysis of chromosome-encoded microcins. Antimicrob. Agents Chemother. 50:1411-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pons, A. M., N. Zorn, D. Vignon, F. Delalande, A. Van Dorsselaer, and G. Cottenceau. 2002. Microcin E492 is an unmodified peptide related in structure to colicin V. Antimicrob. Agents Chemother. 46:229-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas, X., D. Destoumieux-Garzón, J. Péduzzi, C. Afonso, A. Blond, N. Birlirakis, C. Goulard, L. Dubost, R. Thai, J. C. Tabet, and S. Rebuffat. 2004. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 279:28233-28242. [DOI] [PubMed] [Google Scholar]
- 34.Trent, M. S., L. M. Worsham, and M. L. Ernst-Fonberg. 1998. The biochemistry of hemolysin toxin activation: characterization of HlyC, an internal protein acyltransferase. Biochemistry 37:4644-4652. [DOI] [PubMed] [Google Scholar]
- 35.Wallace, B. J., and J. Pittard. 1967. Genetic and biochemical analysis of the isoenzymes concerned in the first reaction of aromatic biosynthesis in Escherichia coli. J. Bacteriol. 93:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh, C. T., J. Liu, F. Rusnak, and M. Sakaitani. 1990. Molecular studies on enzymes in chorismate metabolism and the enterobactin biosynthetic pathway. Chem. Rev. 90:1105-1129. [Google Scholar]
- 37.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 38.Whipp, M. J., and A. J. Pittard. 1995. A reassessment of the relationship between aroK- and aroL-encoded shikimate kinase enzymes of Escherichia coli. J. Bacteriol. 177:1627-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkens, M., J. E. Villanueva, J. Cofre, J. Chnaiderman, and R. Lagos. 1997. Cloning and expression in Escherichia coli of genetic determinants for production of and immunity to microcin E492 from Klebsiella pneumoniae. J. Bacteriol. 179:4789-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reference deleted.
- 41.Zhu, M., M. Valdebenito, G. Winkelmann, and K. Hantke. 2005. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology 151:2363-2372. [DOI] [PubMed] [Google Scholar]