Abstract
Fatty acid synthase in the yeast Cryptococcus neoformans is composed of two subunits encoded by FAS1 and FAS2 genes. We inserted a copper-regulated promoter (PCTR4-2) to regulate FAS1 and FAS2 expression in Cryptococcus neoformans (strains PCTR4-2/FAS1 and PCTR4-2/FAS2, respectively). Both mutants showed growth rates similar to those of the wild type in a low-copper medium in which FAS1 and FAS2 were expressed, but even in the presence of exogenous fatty acids, strains were suppressed in growth under high-copper conditions. The treatment of C. neoformans with fluconazole was shown to have an increased inhibitory activity and even became fungicidal when either FAS1 or FAS2 expression was suppressed. Furthermore, a subinhibitory dose of fluconazole showed anticryptococcal activity in vitro in the presence of cerulenin, a fatty acid synthase inhibitor. In a murine model of pulmonary cryptococcosis, a tissue census of yeast cells in PCTR4-2/FAS2 strain at day 7 of infection was significantly lower than that in mice treated with tetrathiomolybdate, a copper chelator (P < 0.05), and a yeast census of PCTR4-2/FAS1 strain at day 14 of infection in the brain was lower in the presence of more copper. In fact, no positive cultures from the brain were detected in mice (with or without tetrathiomolybdate treatment) infected with the PCTR4-2/FAS2 strain, which implies that this mutant did not reach the brain in mice. We conclude that both FAS1 and FAS2 in C. neoformans are essential for in vitro and in vivo growth in conditions with and without exogenous fatty acids and that FAS1 and FAS2 can potentially be fungicidal targets for C. neoformans with a potential for synergistic behavior with azoles.
Cryptococcus neoformans is a pathogenic basidiomycetous fungus that can cause a lethal meningoencephalitis or pulmonary infection in both immunocompromised patients and apparently immunocompetent patients (1). A number of virulence factors, including high-temperature growth, melanin synthesis, and capsule production, have been found to have a substantial impact on cryptococcal pathogenesis, and these factors and the genes/proteins that control them can potentially be specific targets for antifungal drug development (8). Furthermore, the importance of metabolic functions and energy production for the yeast Cryptococcus neoformans to produce disease is identified in in vivo transcriptional studies of Cryptococcus (14). For example, fatty acids are known to serve many critically important roles in the cell, including energy storage, integrity and dynamics of biological membranes, cellular metabolism, and cellular signaling (16, 18). The de novo type I fatty acid biosynthesis found in eukaryotes involves a complex of highly integrated multifunctional enzymes with various catalytic activities. The type Ib fatty acid synthase system is found in animals and is composed of two identical α subunits (α2), whereas type Ia is found mainly in fungi and some bacteria (16). In Saccharomyces cerevisiae, fatty acid biosynthesis requires acetyl-CoA carboxylase (encoded by the ACC1 gene) and a complex of fatty acid synthases that includes two subunits, α and β, formed as a heterohexamer (α6β6) (16, 22). The trifunctional α subunit is encoded by the FAS2 gene, and the pentafunctional β subunit is encoded by the FAS1 gene (12, 17). The OLE1 gene encodes Δ-9 fatty acid desaturase and is also essential for the production of monounsaturated fatty acids in S. cerevisiae (20) and is probably essential for the survival of C. neoformans since a null mutant could not be created (G. M. Cox, unpublished data). Previous studies of S. cerevisiae have shown that ACC1 is essential for vegetative growth, which infers that ACC1 may have functions other than the biosynthesis of fatty acid (7) since fas1 and fas2 null mutants of S. cerevisiae and Candida albicans are auxotrophic for fatty acids (15, 24, 25). Studies of C. albicans have shown that although fas2 mutants can survive in vitro, they are attenuated for virulence in both murine oropharyngeal candidiasis and candidemia models (24, 25).
In a transcriptional profiling study of C. neoformans for high-temperature growth, the transcriptional activator gene MGA2 was found to be up-regulated for high-temperature growth at 37°C versus 25°C. An mga2 null mutant was created, and in a study of comparative transcriptional profiles (the mga2 mutant versus the wild-type strain), MGA2 was shown to regulate FAS1 and ACC1 expression. The mga2 mutant was also found to be temperature sensitive for growth at 37°C and hypersensitive to the ergosterol synthesis inhibitor fluconazole in vitro (9). These observations allowed us to hypothesize that the fatty acid synthase genes in C. neoformans may be extremely important to membrane structures and functions and a block in fatty acid synthesis could positively affect an azole's impact on ergosterol synthesis and membrane formation.
We made several attempts to disrupt C. neoformans FAS1 and FAS2 homologues by site-directed mutagenesis, and with more than 100 transformants screened, there was no evidence of a homologous recombination event. We predicted that both FAS1 and FAS2 in C. neoformans serve important roles in basic survival and growth of C. neoformans. Therefore, for experimental support of this hypothesis, we regulated the expression of FAS1 and FAS2 by fusing a copper-regulated promoter to replace the FAS1 and FAS2 endogenous promoters and determined the phenotypes of these mutants in vitro and in vivo and with azole exposure. The objective of this study was to examine the potential role of fatty acid synthesis in C. neoformans as a potential fungicidal target.
MATERIALS AND METHODS
Yeast strains, culture media, and additives.
Wild-type C. neoformans strain H99 (serotype A, MATα) was cultured on yeast extract-peptone-dextrose (YPD) medium (1% yeast extract, 2% peptone, and 2% dextrose). Transformants were grown on a YPD plate containing 1 M sorbitol for 3 h and then transferred for selection onto a YPD plate supplemented with 100 μg of nourseothricin (clonNAT; Werner BioAgents, Jena, Germany) per ml and 200 μM bathocuproinedisulfonic acid (BCS; Sigma-Aldrich, St. Louis, MO) as a copper chelator. Subsequent selected transformants with homologous recombination events were grown in YPD medium with either 200 μM BCS or 25 μM cupric sulfate (CuSO4; Sigma-Aldrich, St. Louis, MO). All media with CuSO4 also had additional supplementation with 1 mM ascorbic acid (Sigma-Aldrich, St. Louis, MO). Fatty acid supplementation was achieved by adding 1% (vol/vol) Tween 40 (Sigma-Aldrich, St. Louis, MO), 0.01% (wt/vol) myristic acid (Sigma-Aldrich, St. Louis, MO), and 0.01% (wt/vol) stearic acid (Sigma-Aldrich, St. Louis, MO) to YPD medium.
Identification of FAS1 and FAS2 homologues in C. neoformans.
The C. albicans FAS1 (GenBank accession number X74952) and FAS2 (GenBank accession number L29063) sequences from GenBank were blasted to the C. neoformans strain H99 genomic database (http://cneo.genetics.duke.edu) to identify the putative cryptococcal homologues.
DNA constructs and transformation protocol.
Overlap PCRs were performed to create the pNAT/CTR4-2 plasmid as described previously (4), and oligonucleotides used in this study are shown in Table 1. In brief, a 1.6-kb nourseothricin acetyltransferase dominant selectable marker (Natr) was amplified from plasmid pCH233 by using primers NAT1F and NAT2R. The pCTR4-2/Gust plasmid (a gift from Tamara Doering, Washington University, St. Louis, MO) (13) was used as a template to amplify the 0.7-kb copper-regulated promoter (PCTR4-2) with primers CTR3F and CTR4R. The PCR products were run on a 0.7% agarose gel, stained with ethidium bromide, and gel purified. Primers NAT1F and CTR4R were used for overlap PCR to connect Natr upstream to PCTR4-2, resulting in a 2.2-kb PCR fragment that was cloned into the pCR2.1 vector by using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA). The resulting pNAT/CTR4-2 plasmid was sequenced for confirmation.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Type | Sequence | Reference or source |
|---|---|---|---|
| NAT1F | Forward | 5′-CATGCAGGATTCGAGTGGCATG-3′ | This study |
| NAT2R | Reverse | 5′-ATCGATAAGCTTGATTTGGCGGAGCCATGAAGATCCTGAGGA-3′ | This study |
| CTR3F | Forward | 5′-TCCTCAGGATCTTCATGGCTCCGCCAAATCAAGCTTATCGAT-3′ | This study |
| CTR4R | Reverse | 5′-CATATGGATTGGTGAAGTCG-3′ | This study |
| F1CTR1 | Forward | 5′-AAGGAACCCCGTGTATTTCC-3′ | This study |
| F1CTR2Rev | Reverse | 5′ CACTCGAATCCTGCATGCGACTCTCCTGTGCTGCGG-3′ | This study |
| F1CTR3 | Forward | 5′-CCGCAGCACAGGAGAGTCGCATGCAGGATTCGAGTG-3′ | This study |
| F1CTR4Rev | Reverse | 5′-GAAGAGAAGAGGGAGGCCATGATTGGTGAAGTCGTTGTCG-3′ | This study |
| F1CTR5 | Forward | 5′-CGACAACGACTTCACCAATCATGGCCTCCCTCTTCTCTTC-3′ | This study |
| F1CTR6Rev | Reverse | 5′-CTCGGAGACCGATGTAAAAC-3′ | This study |
| F2CTR1 | Forward | 5′-CGCAAAGGCCTTGAAGTAAG-3′ | This study |
| F2CTR2Rev | Reverse | 5′-CACTCGAATCCTGCATGCTGTGTATGTCTCCGTCGGGG-3′ | This study |
| F2CTR3 | Forward | 5′-CCCCGACGGAGACATACACAGCATGCAGGATTCGAGTG-3′ | This study |
| F2CTR4Rev | Reverse | 5′-CATCCCTTCAGCGGCCATGATTGGTGAAGTCGTTGTCG-3′ | This study |
| F2CTR5 | Forward | 5′-CGACAACGACTTCACCAATCATGGCCGCTGAAGGGATG-3′ | This study |
| F2CTR6Rev | Reverse | 5′-AGCCTTCTTGGCAAAAGTGA-3′ | This study |
| FAS1L-RT | Forward | 5′-ACGCTTCTTACAGCGGTATC-3′ | This study |
| FAS1R-RT | Reverse | 5′-CTCTCTTTGAGCGTGCTCCT-3′ | This study |
| FAS2L-RT | Forward | 5′-TGGTGCTCTCTCTGGTGTTG-3′ | This study |
| FAS2R-RT | Reverse | 5′-GACCAGCAAAGGAGGCAGTA-3′ | This study |
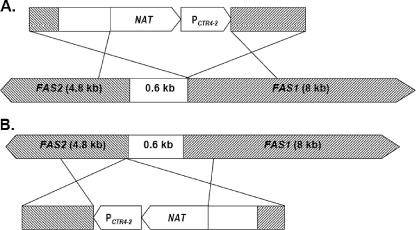
The copper-regulated FAS1 and FAS2 mutants were constructed using the overlap PCR technique. A 990-bp fragment of the 5′ region, including the promoter region of FAS1, was amplified upstream −1 base from the ATG start site with primers F1CTR1 and F2CTR2Rev. A 1,043-bp fragment of the 3′ region of FAS1 was amplified downstream from the ATG start site with primers F1CTR5 and F1CTR6Rev. The 2,280-bp Natr-PCTR4-2 fragment was amplified from the pNAT/CTR4-2 plasmid template by using primers F1CTR3 and F1CTR4Rev. All three PCR products were gel purified and used as templates for overlap PCR by using primers F1CTR1 and F1CTR6Rev, resulting in a 4.2-kb overlap fragment (Fig. 1A). For the copper-regulated FAS2 construct, a 1,079-bp fragment of the 5′ region, including the promoter region of FAS2, was amplified upstream −1 base from the ATG start site with primers F2CTR1 and F2CTR2Rev. A total of 1,114 bp of the 3′ region of FAS1 was amplified downstream from the ATG start site with primers F2CTR5 and F2CTR6Rev. The 2,280-bp Natr-PCTR4-2 fragment was amplified from the pNAT/CTR4-2 plasmid template by using primers F2CTR3 and F2CTR4Rev. All three PCR products were gel purified and used as templates for overlap PCR by using primers F2CTR1 and F2CTR6Rev, resulting in a 4.4-kb overlap fragment (Fig. 1B).
FIG. 1.
Diagram shows FAS1, FAS2, and the putative common promoter in C. neoformans with the genome and the DNA construct fragments. Nourseothricin resistance gene cassette (Natr) and the copper-reguated promoter (PCTR4-2) were inserted by homologous recombination to regulate FAS1 (A) and FAS2 (B).
The overlap DNA constructs generated by overlap PCR were used to transform strain H99 by using a biolistic delivery protocol (21) for homologous recombination into the cryptococcal genome (Fig. 1A and B). Stable transformants were selected on YPD medium containing nourseothricin and BCS as described previously (11). Analyses of the transformants for homologous recombination were performed by PCR of the genomic DNA from transformants with primers flanking the construct. Confirmation of homologous recombination was performed by Southern hybridization. For PCTR4-2/FAS1 mutants, genomic DNA was digested individually with either KpnI or ClaI and probed with a [32P]dCTP-labeled FAS1 3′ fragment, whereas the genomic DNA of PCTR4-2/FAS2 was digested individually with KpnI, EcoRI, and HindIII and probed with a [32P]dCTP-labeled FAS2 5′ fragment.
Determination of FAS1 and FAS2 mRNA by quantitative RT-PCR.
C. neoformans strains H99, PCTR4-2/FAS1, and PCTR4-2/FAS2, were grown at 30°C in two flasks each of 50 ml YPD medium containing BCS to mid-log phase. The cells were pelleted, washed twice with sterile phosphate-buffered saline (PBS), and resuspended in 50 ml YPD medium containing either BCS or CuSO4 for 6 h. The cells were then pelleted, frozen at −80°C, and lyophilized. The lyophilized cells were broken with 2-mm glass beads, and RNA was extracted from these strains by using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. One microgram of RNA was treated with DNase I, followed by cDNA synthesis using oligo(dT) primers with the SuperScript III first-strand synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen Life Technologies, Carlsbad, CA). The resulting cDNA was diluted 1:5, and 2 μl of diluted cDNA was used as the template for quantitative RT-PCR using iQ SYBR green Supermix (Bio-Rad, Hercules, CA). Real-time PCR primer sequences for FAS1 (FAS1L-RT and FAS1R-RT) and FAS2 (FAS2L-RT and FAS2R-RT) were added to the reaction mixtures at a final concentration of 0.1 μM. The iCycler iQ multicolor real-time detection system (Bio-Rad, Hercules, CA) was used with the following conditions: an initial denaturing cycle of 95°C for 3 min, 40 cycles of denaturation at 95°C for 20 s, and annealing/extension at 60°C for 45 s, followed by a melt curve from 60°C to 95°C with fluorescent monitoring for each 0.5°C.
Determination of yeast cell growth.
C. neoformans strains H99, PCTR4-2/FAS1, and PCTR4-2/FAS2 were grown at 30°C in YPD broth or in YPD broth containing BCS. Cells were pelleted, washed twice with sterile PBS, and resuspended in PBS with 1:10 serial dilutions for six dilutions. A total of 50 μl each of diluted yeast cells (106 to 10 cells) was placed onto YPD agar and YPD agar containing BCS or CuSO4 with and without fatty acid supplementation. Cryptococcal strains were incubated at both 30°C and 37°C. Growth curves were created for 105 cells of C. neoformans strains. H99, PCTR4-2/FAS1, and PCTR4-2/FAS2 strains were cultured at 30°C and 37°C in YPD broth containing either BCS or CuSO4 with and without fatty acid supplementation. Quantitative yeast cell counts were performed by culture of 100 μl of diluted yeast cells onto YPD-BCS agar. Quantitative counts by CFU on YPD-BCS agar were determined at 0, 6, 12, 24, 48, and 72 h after culture.
Determination of synergistic activity of antifungal agents with FAS inhibition.
A total of 1 × 105 cells of C. neoformans strains H99, PCTR4-2/FAS1, and PCTR4-2/FAS2 were grown at 30°C in YPD broth containing BCS or CuSO4 with and without either 8 μg/ml fluconazole or 4 μg/ml caspofungin. Quantitative cell counts were performed by plating 100-μl aliquots of diluted yeast cells on YPD-BCS agar for CFU at 0, 6, 12, 24, 48, and 72 h of exposure. Then, 1 × 105 yeast cells of C. neoformans strain H99 were grown at 30°C in YPD broth with 8 μg/ml fluconazole, 0.3 μg/ml cerulenin, and both together. Quantitative yeast cell counts were performed by culture of 100 μl of diluted yeast cells on YPD agar. Quantitative counts by CFU on YPD agar were determined at 0, 6, 12, 24, and 48 h of culture.
In vivo regulation of copper-regulated FAS1 and FAS2 expression in a murine model.
A murine model of pulmonary cryptococcosis was used to determine the importance of FAS expression by in vivo copper-regulated FAS1 and FAS2 expression mutants. A total of 1 × 105 yeast cells of C. neoformans strains H99, PCTR4-2/FAS1, and PCTR4-2/FAS2 were inoculated intranasally into A/JCr mice. Thirty mice were used for each group, and 15 mice of each group were treated with 0.2 μg/day of ammonium tetrathiomolybdate (TM), a copper chelator, daily and fed a low-copper diet (5). In the TM group, TM was injected intraperitoneally into mice for 2 weeks before inoculation to ensure a low-copper condition and daily injection continued throughout the experiment. The control groups were given a regular diet and water. After 14 days of TM treatment, the ceruloplasmin concentration in these mice had been reduced 25% compared to the concentration in control mice. Five mice from each group were sacrificed, and quantitative yeast cultures from murine lungs and brains were performed at 4, 7, and 14 days after inoculation. Statistical analyses were performed using the Student t test to calculate a P value for the tissue census of groups, and a P value of ≤0.05 was considered significant.
RESULTS
Identification and regulation of FAS1 and FAS2 in C. neoformans.
The C. albicans FAS1 and FAS2 DNA sequence was used to search for their homologues in the C. neoformans genome. In the genome database of C. neoformans var. grubii (serotype A) strain H99 (Broad Institute; http://www.broad.mit.edu), FAS1 and FAS2 were found closely linked in opposite orientations and were arranged around an apparent 0.6-kb putative common promoter (Fig. 1). In C. neoformans strain H99, the FAS1 gene was 8,060 bp and contained nine introns; the putative amino acid sequence of FAS1 was 2,514 amino acids (GenBank accession number DQ363334). The FAS2 gene was 4,829 bp and contained nine introns; the predicted amino acid sequence of FAS2 was 441 amino acids (GenBank accession number DQ363335). The selection of PCTR4-2/FAS1 and PCTR4-2/FAS2 mutants was performed by PCR using primers that flank the construct and by Southern hybridization (see Materials and Methods). Our results confirmed homologous recombination and replacement of the wild-type gene with the copper-regulated promoters in our mutant strains PCTR4-2/FAS1 and PCTR4-2/FAS2 (data not shown).
Determination of FAS1 and FAS2 mRNA by quantitative real-time PCR.
Real-time PCR was performed to quantitate the FAS1 and FAS2 transcripts of C. neoformans strains H99, PCTR4-2/FAS1, and PCTR4-2/FAS2 in CuSO4 conditions compared to those in BCS conditions. Neither BCS nor CuSO4 conditions showed significant impact on FAS1 and FAS2 transcripts of wild-type parental C. neoformans strain H99. In the PCTR4-2/FAS1 strain, BCS and CuSO4 exposure showed little impact on FAS2 transcripts but the FAS1 transcript had a 200-fold reduction in CuSO4 compared to in BCS. In contrast to the PCTR4-2/FAS1 strain, BCS and CuSO4 showed no significant impact on FAS1 transcripts in the PCTR4-2/FAS2 strain, whereas a 500-fold reduction of FAS2 transcripts was observed in the PCTR4-2/FAS2 strain with CuSO4 compared to the transcript level with BCS. This demonstrates that the PCTR4-2/FAS2 strain is even more sensitive to copper exposure in its expression of FAS2 than the PCTR4-2/FAS1 strain is with FAS1.
Determination of cell growth and growth curves.
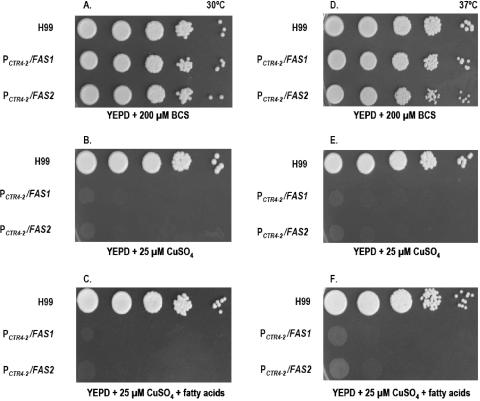
Cell growth was determined in YPD agar with BCS, CuSO4, or CuSO4 plus fatty acids at 30°C and 37°C as shown in Fig. 2A. At both 30°C and 37°C, C. neoformans strain H99 shows normal growth on YPD agar containing BCS, YPD agar containing CuSO4, and YPD agar containing CuSO4 with fatty acids. In contrast, both mutant strains show a complete growth arrest on YPD agar containing CuSO4 acid and on YPD agar containing CuSO4 with fatty acid supplementation at both temperatures.
FIG. 2.
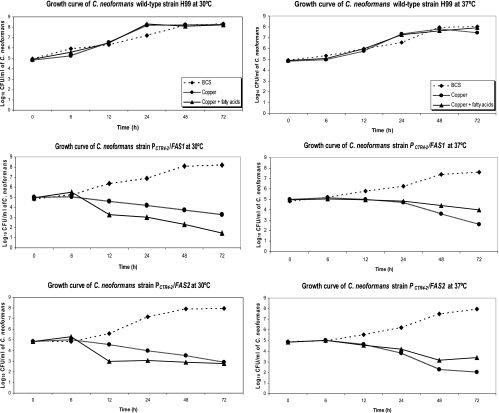
(A) Growth determination of C. neoformans strains H99, PCTR4-2/FAS1, and PCTR4-2/FAS2 on YPD agar containing BCS (A and D), CuSO4 (B and E), and CuSO4 with fatty acids (C and F) at 30°C and 37°C (serial 10-fold dilutions from left to right). (B) Growth curves of C. neoformans wild-type strain H99, PCTR4-2/FAS1, and PCTR4-2/FAS2 in YPD broth containing BCS, CuSO4, and CuSO4, with fatty acids at 30°C and 37°C.
Growth curves were determined for H99, PCTR4-2/FAS1, and PCTR4-2/FAS2 strains (Fig. 2B). A total of 1 × 105 cells of each strain of C. neoformans were grown at both 30°C and 37°C in YPD broth containing BCS or CuSO4 with and without fatty acid supplementation. With H99, the yeast cells showed equal growth levels in media containing BCS, CuSO4, or CuSO4 with fatty acids in both temperatures, and saturated conditions were achieved at 24 h. In contrast, after 6 h in culture, the yeast cell numbers of the PCTR4-2/FAS1 and PCTR4-2/FAS2 strains began to decrease to concentrations approximately 100-fold less than the starting yeast cell numbers at 72 h in CuSO4.
Both PCTR4-2/FAS1 and PCTR4-2/FAS2 strains represented approximately the same growth rate as H99 in YPD with BCS in both 30°C and 37°C and also exhibited a normal capsule, cell morphology by microscopy, and melanin production on 3,4-dihydroxyphenyalanine plates.
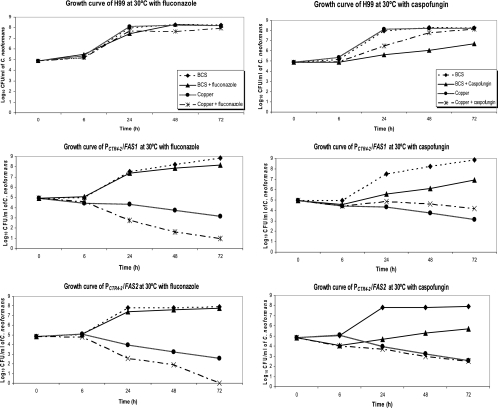
We next examined the antifungal activity of fluconazole and caspofungin in conditions in which either FAS1 or FAS2 was inhibited. A total of 1 × 105 wild-type H99, PCTR4-2/FAS1, and PCTR4-2/FAS2 cells were grown at 30°C in YPD broth containing either BCS or CuSO4. Then, fluconazole or caspofungin was added into the growth media with BCS and CuSO4, and growth curves were determined. Fluconazole at 8 μg/ml had no significant impact on cell growth of H99 in BCS or CuSO4 (subinhibitory dose). In BCS, the yeast cell counts of both PCTR4-2/FAS1 and PCTR4-2/FAS2 decreased about only 0.5 log when fluconazole was added. On the other hand, fluconazole, a cell membrane inhibitor enzyme, possessed more antifungal activity with PCTR4-2/FAS1 with CuSO4 exposure in which FAS1 was inhibited. The viable yeast cells were 100-fold less than the number with only CuSO4 at 72 h of culture incubation. Similarly, fluconazole also had more antifungal activity with strain PCTR4-2/FAS2 in CuSO4 than with the other strains and, actually, fluconazole became fungicidal in CuSO4 at 72 h, with the death of the entire inoculum (Fig. 3).
FIG. 3.
Growth curves for antifungal activity of fluconazole (8 μg/ml) and caspofungin (4 μg/ml) to H99, PCTR4-2/FAS1, and PCTR4-2/FAS2 in conditions with BCS or CuSO4 at 30°C.
In contrast, caspofungin (a cell wall enzyme inhibitor) at 4 μg/ml had little anticryptococcal activity and there was no detectable increase in antifungal activity for the three strains in all conditions. For example, when strains were grown in BCS or CuSO4 in the presence or absence of caspofungin, the reductions in CFU with and without caspofungin are not different (Fig. 3).
Interaction of cerulenin and fluconazole on C. neoformans.
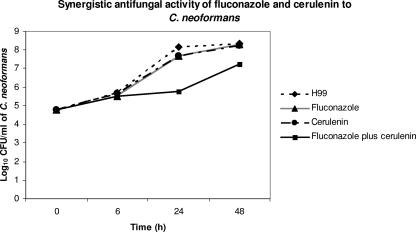
H99 was used to determine the interaction between the antifungal activity of fluconazole and a known fatty acid synthase inhibitor, cerulenin. Fluconazole at 8 μg/ml, cerulenin at 0.3 μg/ml, or both together were added to 1 × 105 cells of C. neoformans in YPD broth at 30°C. Quantitative yeast counts showed that medium with a combination of cerulenin and fluconazole had more inhibition of growth than medium with either drug alone at 24 and 48 h (Fig. 4).
FIG. 4.
Growth curves for single and combined antifungal activity of fluconazole (8 μg/ml) and cerulenin (0.3 μg/ml) to H99 at 30°C.
In vivo regulation of copper-regulated FAS1 and FAS2 strains in a murine model.
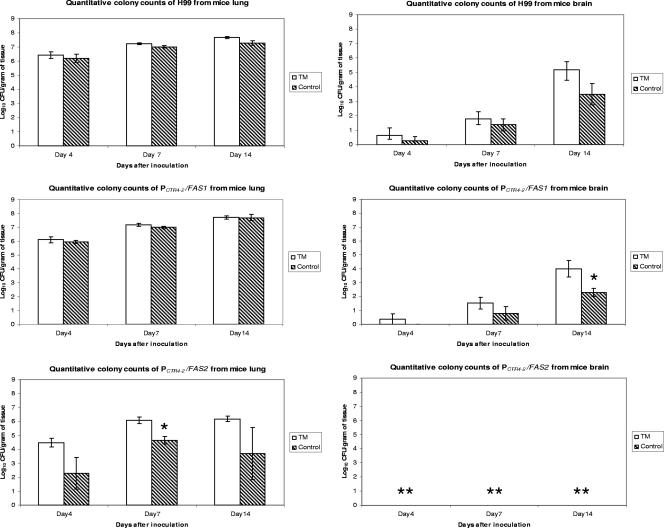
We examined the regulation of FAS1 and FAS2 under variable copper exposure in a murine model of pulmonary cryptococcosis using PCTR4-2/FAS1 and PCTR4-2/FAS2 strains of C. neoformans. TM was used as a copper chelator in mice and was able to reduce tissue copper levels, as reflected by lower ceruloplasma levels in treated animals. In mice infected with H99 and PCTR4-2/FAS1, the tissue census recovered from the lungs was slightly lower in the control group than in the TM-treated group but it was not statistically significant. However, the tissue yeast counts recovered from murine lung infected with the PCTR4-2/FAS2 strain were consistently lower at 4, 7, and 14 days in the control group than in the TM-treated group and, at day 7, yeast counts were significantly reduced (P < 0.05) (Fig. 5). The yeast counts from brains of animals infected with H99 were not significantly different in the control versus the TM-treated group of animals. On the other hand, compared to the result for the TM-treated group, murine brains infected with PCTR4-2/FAS1 showed fewer yeast colony counts than control brains did and, at day 14 of infection, these yeast counts were significantly reduced (P < 0.05). Furthermore, no positive cultures for the PCTR4-2/FAS2 strain were found from murine brain in either control or TM-treated animals and, thus, this mutant neither reached nor survived in the brain.
FIG. 5.
Quantitative CFU from murine lung and brain of H99, PCTR4-2/FAS1, and PCTR4-2/FAS2 in murine model of pulmonary cryptococcosis at days 4, 7, and 14 of infection. *, statistical significance (P < 0.05); **, no positive cultures from brain. Error bars indicate standard deviations.
DISCUSSION
Lipid metabolism in eukaryotes, including the biosynthesis of saturated long-chain fatty acids, is vitally important to cell integrity, signaling, and energy production. Thus, fatty acid synthesis has been a promising area for antifungal drug development, and the genes related to the initial fatty acid biosynthesis may serve as specific antifungal targets. The present study supports the fact that both FAS1 and FAS2 are essential for vegetative survival of C. neoformans. Furthermore, when either FAS1 or FAS2 expression is suppressed, C. neoformans is not able to grow and fungal death occurs in culture during FAS suppression even in the presence of exogenous fatty acids in the medium. These findings are different from those observed with S. cerevisiae and C. albicans in which the fas1 and fas2 null mutants are auxotrophic for fatty acids. A possible explanation for these divergent findings is that FAS1 and FAS2 in C. neoformans may have other functions besides direct fatty acid metabolism or that C. neoformans does not have a mechanism to transport and/or activate exogenous fatty acids into the cell through its capsule, cell wall, or cell membrane. Consequently, with the deficiency of intracellular fatty acids, the yeast is unable to effectively produce its biological membranes, which are essential for cell division and/or cell metabolism, and, therefore, death occurs rapidly. Fatty acid transport in C. neoformans has not yet been characterized. In S. cerevisiae, the fatty acyl-CoA synthetase (Faa1 and Faa4) and Fat1 have been shown to be responsible for fatty acid import by linking transport and activation of exogenous fatty acids for utilization and signaling in the cell (6, 22). We blasted the FAA1 and FAT1 genes of S. cerevisiae into the C. neoformans genome database, and no similar genes were found. Thus, C. neoformans may have a different system of fatty acid transport/activation, and this possibility needs further study.
We found that the FAS1 and FAS2 genes are closely linked on the same chromosome, with opposite orientations, which is different from that of FAS1 and FAS2 in C. albicans, which are located on different chromosomes. The 8-kb FAS1 and 4.8-kb FAS2 are controlled by a relatively small putative common promoter. We inserted the Natr-PCTR4-2 fragment to replace the native promoters of the FAS1 or FAS2 open reading frames. Quantitative RT-PCR demonstrated that the mRNA of FAS1 or FAS2 did not significantly change when the Natr-PCTR4-2 fragment was inserted upstream to FAS2 or FAS1, respectively. Therefore, this replacement promoter was efficient in the control of expression of only FAS1 or FAS2 in PCTR4-2/FAS1 and PCTR4-2/FAS2 mutants, respectively. On the other hand, the quantitative RT-PCR results did show that the mRNAs of FAS1 or FAS2 were significantly down-regulated in the PCTR4-2/FAS1 and PCTR4-2/FAS2 strains in the presence of copper ion. In the comparison of the two engineered strains, PCTRA-2/FAS2 appears to be more sensitive to exogenous copper effects than PCTRA-2/FAS1 is and this sensitivity might have relevance for in vivo environments.
Since fatty acids are important components of lipid membranes, synergistic activity between fatty acid and other membrane synthesis inhibitors was predicted. In fact, in our microarray studies, the transcriptional activator MGA2 was shown to be linked to the control of fatty acid synthesis gene expression and cell membrane phenotype when the mga2 mutant was found to be hypersensitive to fluconazole exposure (9). In the present study, we directly show that when fatty acid synthase is inhibited, fluconazole has more potent direct antifungal activity against C. neoformans and, in fact, fluconazole becomes fungicidal in vitro. For example, when FAS1 or FAS2 expression is down-regulated, the number of surviving C. neoformans CFU decreases approximately 100-fold more with fluconazole exposure and death ensues. In contrast, the cell wall inhibitor caspofungin does not show a synergistic effect with FAS1 or FAS2 inhibition. However, both BCS and CuSO4 enhance the limited anticryptococcal activity of caspofungin. In order to show the proof of concept that these effects could also be reproduced by external sources, we examined the interaction of fluconazole with a known fatty acid synthase inhibitor, cerulenin. The combination of fluconazole and cerulenin showed an approximately 100-fold decrease in yeast cell growth of H99 compared to the result for exposure with the same concentration of fluconazole or cerulenin used alone at 24 h, and a 10-fold decrease persisted at 48 h. These drug combination experiments confirmed that a two-pronged attack on the cell membrane by block of both ergosterol and fatty acid synthesis would be additive to the growth inhibition of C. neoformans.
We next attempted to show that fatty acid synthesis in C. neoformans is important within the mammalian host. We have developed a tetracycline-regulated system (2), but it has not yet been made efficient for in vivo studies and, therefore, we used the copper-promoter system for gene regulation in vivo. The CTR4 gene putatively encodes a high-affinity copper transporter in C. neoformans, and this is similar to its function in Schizosaccharomyces pombe. Recent work has shown the potential importance of copper for cryptococcal growth in the central nervous system. For instance, the copper transcriptional factor, a cuf1 mutant, cannot grow in the brains of mice (23). Also, the CTR4 gene has been shown to be up-regulated in the cerebrospinal fluid of rabbits with cryptococcal meningitis (19). Therefore, in our animal studies, it was important to observe the impact of the in vivo copper conditions we created and whether they had a direct effect on the virulence composite of wild-type C. neoformans H99 through influence on the native CTR4 and CUF1 genes. In fact, the H99 (wild-type) strain grew well and normally in the murine brain with or without the use of TM chelator and this supports the fact that a possible endogenous impact of copper levels on normal genes in this model was not primarily responsible for the effects observed in the fas mutants. On the other hand, our in vivo experiment to regulate FAS2 by copper has shown that a copper-regulated promoter can potentially be used to control the gene expression of C. neoformans in a murine model of pulmonary cryptococcosis. Our in vitro quantitative expression assessment showed that the PCTR4-2/FAS2 was more tightly regulated by copper than the PCTR4-2/FAS1 was. This feature may also have been observed in vivo since compared to PCTR4-2/FAS1 only the PCTR4-2/FAS2 strain showed a significant decrease in CFU counts in the lung at day 7 after infection in the presence of normal host copper levels compared to animals that had reduced available copper through chelation. It is difficult to chelate all copper out of mice since there are side effects of the chelator at high concentrations, and complete elimination of copper from the host is a limitation of this strategy to precisely cut off gene expression. However, ceruloplasmin levels by our treatment were decreased by 25% and this finding is similar to those of other studies which have used this method (3, 5). Furthermore, it was also shown that an altered promoter for FAS1 and FAS2 can have significant impact on yeast entry or survival in the brain. The colony counts of the PCTR4-2/FAS1 strain revealed a significant decrease in brain yeast counts 2 weeks after infection in the presence of normal host copper, whereas no yeast cells from the PCTR4-2/FAS2 strain were able to grow in murine brain after intranasal infection in both low-copper and normal copper conditions. These observations suggest that the mutants with regulated FAS genes have difficulty either surviving in the lung and/or escaping from the lung into the bloodstream and entering the brain. It is apparent from these in vivo experiments that any inhibition of fatty acid synthesis can potentially cause a substantial impact on the yeast's ability to survive in vivo.
Human fatty acid synthase encoded by the FASN gene is a type Ib fatty acid synthase. Thus, there is a possibility that the yeast type Ia fatty acid synthase can potentially be used as a unique fungicidal target for C. neoformans, and the target shows some breadth in that it is important for in vivo growth of C. albicans (24, 25). Furthermore, there are a number of fatty acid synthase inhibitors isolated from plants that may have antifungal activity (10). Further study into the function(s) of fatty acid synthases in C. neoformans and into identifying compounds which have inhibitory activity for fatty acid synthases are is promising as a novel antifungal target(s) for drug development both as individual drugs and, particularly, in combination with established cell membrane antifungal agents like the azoles.
Acknowledgments
We acknowledge Tamara Doering (Washington University, St. Louis, MO) for providing pCTR4-2/Gust plasmid, Joanne Kingsbury (Duke University Medical Center, Durham, NC) for her advice about the materials and methods in experiments related to copper-regulated promoter, Gary M. Cox (Duke University Medical Center, Durham, NC) for his advice about the fatty acid synthesis pathway, Kirsten Nielsen (Duke University Medical Center, Durham, NC) for assistance in animal study, and Wiley A. Schell and Jackie L. Miller (Duke University Medical Center, Durham, NC) for their advice in antimicrobial susceptibility testing.
This work was supported by Public Health Service grant AI28388.
Footnotes
Published ahead of print on 13 August 2007.
REFERENCES
- 1.Chayakulkeeree, M., and J. R. Perfect. 2006. Cryptococcosis. Infect. Dis. Clin. N. Am. 20:507-544. [DOI] [PubMed] [Google Scholar]
- 2.Chayakulkeeree, M., and J. R. Perfect. 2005. Abstr. 6th Int. Conf. Cryptococcus Cryptococcosis, abstr. MB5.
- 3.Cox, C., T. N. Teknos, M. Barrios, G. J. Brewer, R. D. Dick, and S. D. Merajver. 2001. The role of copper suppression as an antiangiogenic strategy in head and neck squamous cell carcinoma. Laryngoscope 111:696-701. [DOI] [PubMed] [Google Scholar]
- 4.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 5.Elner, S. G., V. M. Elner, A. Yoshida, R. D. Dick, and G. J. Brewer. 2005. Effects of tetrathiomolybdate in a mouse model of retinal neovascularization. Investig. Ophthalmol. Visual Sci. 46:299-303. [DOI] [PubMed] [Google Scholar]
- 6.Faergeman, N. J., P. N. Black, X. D. Zhao, J. Knudsen, and C. C. DiRusso. 2001. The Acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular utilization. J. Biol. Chem. 276:37051-37059. [DOI] [PubMed] [Google Scholar]
- 7.Hasslacher, M., A. S. Ivessa, F. Paltauf, and S. D. Kohlwein. 1993. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J. Biol. Chem. 268:10946-10952. [PubMed] [Google Scholar]
- 8.Idnurm, A., Y. S. Bahn, K. Nielsen, X. Lin, J. A. Fraser, and J. Heitman. 2005. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3:753-764. [DOI] [PubMed] [Google Scholar]
- 9.Kraus, P. R., M. J. Boily, S. S. Giles, J. E. Stajich, A. Allen, G. M. Cox, F. S. Dietrich, J. R. Perfect, and J. Heitman. 2004. Identification of Cryptococcus neoformans temperature-regulated genes with a genomic-DNA microarray. Eukaryot. Cell 3:1249-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, X. C., A. S. Joshi, H. N. ElSohly, S. I. Khan, M. R. Jacob, Z. Zhang, I. A. Khan, D. Ferreira, L. A. Walker, S. E. Broedel, Jr., R. E. Raulli, and R. L. Cihlar. 2002. Fatty acid synthase inhibitors from plants: isolation, structure elucidation, and SAR studies. J. Nat. Prod. 65:1909-1914. [DOI] [PubMed] [Google Scholar]
- 11.Missall, T. A., and J. K. Lodge. 2005. Thioredoxin reductase is essential for viability in the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 4:487-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohamed, A. H., S. S. Chirala, N. H. Mody, W. Y. Huang, and S. J. Wakil. 1988. Primary structure of the multifunctional alpha subunit protein of yeast fatty acid synthase derived from FAS2 gene sequence. J. Biol. Chem. 263:12315-12325. [PubMed] [Google Scholar]
- 13.Ory, J. J., C. L. Griffith, and T. L. Doering. 2004. An efficiently regulated promoter system for Cryptococcus neoformans utilizing the CTR4 promoter. Yeast 21:919-926. [DOI] [PubMed] [Google Scholar]
- 14.Rude, T. H., D. L. Toffaletti, G. M. Cox, and J. R. Perfect. 2002. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect. Immun. 70:5684-5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schüller, H. J., B. Fortsch, B. Rautenstrauss, D. H. Wolf, and E. Schweizer. 1992. Differential proteolytic sensitivity of yeast fatty acid synthetase subunits alpha and beta contributing to a balanced ratio of both fatty acid synthetase components. Eur. J. Biochem. 203:607-614. [DOI] [PubMed] [Google Scholar]
- 16.Schweizer, E., and J. Hofmann. 2004. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol. Mol. Biol. Rev. 68:501-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schweizer, M., L. M. Roberts, H. J. Holtke, K. Takabayashi, E. Hollerer, B. Hoffmann, G. Muller, H. Kottig, and E. Schweizer. 1986. The pentafunctional FAS1 gene of yeast: its nucleotide sequence and order of the catalytic domains. Mol. Gen. Genet. 203:479-486. [DOI] [PubMed] [Google Scholar]
- 18.Shea, J. M., J. L. Henry, and M. Del Poeta. 2006. Lipid metabolism in Cryptococcus neoformans. FEMS Yeast Res. 6:469-479. [DOI] [PubMed] [Google Scholar]
- 19.Steen, B. R., S. Zuyderduyn, D. L. Toffaletti, M. Marra, S. J. Jones, J. R. Perfect, and J. Kronstad. 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot. Cell 2:1336-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stukey, J. E., V. M. McDonough, and C. E. Martin. 1989. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 264:16537-16544. [PubMed] [Google Scholar]
- 21.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trotter, P. J. 2001. The genetics of fatty acid metabolism in Saccharomyces cerevisiae. Annu. Rev. Nutr. 21:97-119. [DOI] [PubMed] [Google Scholar]
- 23.Waterman, S. R., M. Hacham, G. Hu, X. Zhu, Y. D. Park, S. Shin, J. Panepinto, T. Valyi-Nagy, C. Beam, S. Husain, N. Singh, and P. R. Williamson. 2007. Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J. Clin. Investig. 117:794-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao, X. J., G. E. McElhaney-Feser, W. H. Bowen, M. F. Cole, S. E. Broedel, Jr., and R. L. Cihlar. 1996. Requirement for the Candida albicans FAS2 gene for infection in a rat model of oropharyngeal candidiasis. Microbiology 142:2509-2514. [DOI] [PubMed] [Google Scholar]
- 25.Zhao, X. J., G. E. McElhaney-Feser, M. J. Sheridan, S. E. Broedel, Jr., and R. L. Cihlar. 1997. Avirulence of Candida albicans FAS2 mutants in a mouse model of systemic candidiasis. Infect. Immun. 65:829-832. [DOI] [PMC free article] [PubMed] [Google Scholar]