Abstract
In this study we investigated the interplay of antibiotic pharmacokinetic profiles and the development of mutation-mediated resistance in wild-type and hypermutable Pseudomonas aeruginosa strains. We used in vitro models simulating profiles of the commonly used therapeutic drugs meropenem and ceftazidime, two agents with high levels of antipseudomonal activity said to have different potentials for stimulating resistance development. During ceftazidime treatment of the wild-type strain (PAO1), fully resistant mutants overproducing AmpC were selected rapidly and they completely replaced wild-type cells in the population. During treatment with meropenem, mutants of PAO1 were not selected as rapidly and showed only intermediate resistance due to the loss of OprD. These mutants also replaced the parent strain in the population. During the treatment of the mutator P. aeruginosa strain with meropenem, the slowly selected mutants did not accumulate several resistance mechanisms but only lost OprD and did not completely replace the parent strain in the population. Our results indicate that the commonly used dosing regimens for meropenem and ceftazidime cannot avoid the selection of mutants of wild-type and hypermutable P. aeruginosa strains. For the treatment outcome, including the prevention of resistance development, it would be beneficial for the antibiotic concentration to remain above the mutant prevention concentration for a longer period of time than it does in present regimens.
Pseudomonas aeruginosa is one of the major causes of acute nosocomial infections (34, 36) and the most common pathogen associated with morbidity and mortality among patients with cystic fibrosis and other chronic lung conditions (10, 11, 22).
Resistance development in P. aeruginosa due to the selection of mutants during antimicrobial therapy is a frequent and serious problem. This is especially true for cystic fibrosis patients with chronic lung infections, in whom hypermutable P. aeruginosa strains are found at high frequencies (5, 23, 28). These strains show an increased spontaneous mutation rate due mostly to defects in the DNA methyl-directed mismatch repair system (23, 29). The hypermutation phenotype seems to be an advantage for adaptation to a heterogeneous and fluctuating environment like the lung of a chronically infected patient (28) and is the main cause of the development of multidrug resistance (23, 30).
Antibiotics with high levels of antibacterial activity and low potentials for stimulating resistance development are ideal agents for the treatment of P. aeruginosa infections. Both the β-lactams meropenem and ceftazidime show a high degree of antipseudomonal activity. However, in contrast to ceftazidime, meropenem is said to have low potential for stimulating resistance development (7).
In connection with mutant enrichment, the mutant selection window is generally discussed (9). The mutant selection window is defined as the range of concentrations of an antimicrobial agent extending from the minimum concentration that blocks the growth of the majority of wild-type cells up to that required to block the growth of the least susceptible one-step mutant of the bacterial strain. The lower concentration is close to the MIC of the antimicrobial drug. The upper boundary of the mutant selection window is also called the mutant prevention concentration (MPC) (9). According to this paradigm, due to the lack of a selective advantage, enrichment with resistant mutants does not occur when the drug concentration is below the MIC for the parent strain during a dosing interval.
The most prevalent mutation-mediated mechanism of resistance to meropenem is the loss of the porin OprD (31). Reduced production of OprD in so-called nfxC-type mutants confers slightly reduced susceptibility to meropenem (27). Furthermore, meropenem is a substrate of the MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps, and their upregulated production causes a decrease in meropenem susceptibility (25). Mutation-mediated resistance to ceftazidime is, in the majority of cases, due to the overproduction of the chromosomally encoded inducible AmpC β-lactamase (18). The inactivation of the amidase AmpD leads to an increase of inducer molecules and was found to be the most prevalent mechanism of AmpC hyperproduction in clinical strains (14). Recently, a three-step escalating mechanism of ampC upregulation due to mutations in ampD and two additional ampD homologues, ampDh2 and ampDh3, was elucidated (15). Furthermore, the hyperproduction of the efflux pump MexAB-OprM leads to reduced ceftazidime susceptibility (25).
In the past, several in vitro studies were conducted to investigate and improve the efficacies of meropenem (3, 35) and ceftazidime (1, 26) treatments against P. aeruginosa infections. Enrichment with meropenem- and ceftazidime-resistant mutants of wild-type P. aeruginosa PAO1 and a hypermutable variant of P. aeruginosa PAO1 during exposure to constant antimicrobial concentrations has been studied previously (30). However, to our knowledge, no previous studies have investigated hypermutable P. aeruginosa strains under the influence of clinically achievable pharmacokinetic profiles of meropenem and ceftazidime. Moreover, no elaborated quantitative, phenotypical, and genotypical characterization of the mutants selected has been carried out, and the hypothesis of mutant selection windows has not been taken into consideration to investigate the emergence of resistance.
The aim of our study was to address these issues. Therefore, we used in vitro models simulating the present standard dosing regimens for meropenem and ceftazidime to examine the P. aeruginosa wild-type strain PAO1 and a clinical hypermutable P. aeruginosa strain.
MATERIALS AND METHODS
Bacterial strains and test medium.
P. aeruginosa wild-type strain PAO1 and a clinical, hypermutable, multidrug-resistant, meropenem-susceptible strain, P. aeruginosa 12-09-15, were used for all in vitro simulations. P. aeruginosa 12-09-15 was isolated from a wound swab from a 63-year-old patient in an intensive care unit of a German hospital in 2004. All experiments were performed with Mueller-Hinton medium (BBL Mueller-Hinton II broth, cation adjusted; Becton Dickinson and Company, Sparks, MD).
Susceptibility tests.
Antimicrobial susceptibility testing was performed using the broth microdilution method recommended by the CLSI (6).
Mutation frequency determination.
For P. aeruginosa strain 12-09-15, the mutation frequency on selective rifampin (300 mg/liter) agar was determined in triplicate as described elsewhere (29).
In vitro model experiments.
Ceftazidime (GlaxoSmithKline) and meropenem (AstraZeneca) were kindly provided by the manufacturers. We simulated concentration-time profiles of meropenem and ceftazidime during a dosing regimen of three-times-daily (dosing interval, 8 h) short-time infusions of 1 and 2 g, respectively, by using pharmacokinetic data published by Krueger et al. and Luethy et al. (17, 20). The pharmacokinetic parameters were as follows. For meropenem, the maximum concentration of the drug (Cmax) was 56.1 mg/liter and the half-life was 0.45 h, and for ceftazidime, the Cmax was 173.8 mg/liter and the half-life was 1.96 h. For the simulation of concentration-time curves, the in vitro model of Grasso et al. (12) with slight variations was used. The central compartment (150 ml) of the model was inoculated with an overnight culture of the respective bacterial test strain. After 1 h of incubation at 37°C, an exponential-phase culture was obtained and treatment was started with an initial inoculum of approximately 2.1 × 107 ± 0.686 × 107 CFU/ml. Counts of bacterial colonies were determined and corrected for the loss of CFU due to dilution according to the method of Keil and Wiedemann (16). Mutants were detected on selective media (ceftazidime at 8 mg/liter for P. aeruginosa PAO1 and meropenem at 2 and 8 mg/liter for P. aeruginosa PAO1 and P. aeruginosa 12-09-15, respectively). The detection limit for wild-type and mutant colonies was 1 CFU/ml.
Determination of antibiotic concentration.
Samples taken during the in vitro model experiments were additionally investigated for antibiotic concentrations by a microbiological assay according to a previously described protocol (16) using Klebsiella pneumoniae IV-02-03 and Klebsiella oxytoca IV-03-61 as test organisms for meropenem and ceftazidime, respectively.
Antibacterial effect and pharacokinetic-pharmacodynamic analysis.
The difference between the lowest log10 number of CFU per millimeter observed in a killing curve and the log10 number of CFU per milliliter of the initial inoculum indicated the maximal bactericidal effect (Rmax). Determining the areas above the killing curves (AACs) facilitated the comparison of killing curves. AACs for all killing curves from 0 to 24 h were calculated by the trapezoidal method as previously described (32). By using the pharmacokinetic data for meropenem and ceftazidime published by Krueger et al. and Luethy et al. (17, 20), the cumulative percentage of the dosing interval during which the drug concentration exceeded the MIC, T>MIC, as well as the Cmax/MIC ratio, was determined for each simulation.
Efflux pump inhibition test.
Efflux pump overexpression was detected using an efflux pump inhibitor test (19). Levofloxacin MICs were tested either with or without a 20-μg/ml concentration of the efflux pump inhibitor Phe-Arg-β-naphthylamide (Sigma-Aldrich, Taufkirchen, Germany). A reduction of the levofloxacin MIC by at least 32-fold in the presence of Phe-Arg-β-naphthylamide demonstrated the overproduction of efflux pumps.
Cephalosporinase inhibition test.
The overproduction of the chromosomally encoded cephalosporinase AmpC was evaluated by a disk diffusion test with 30-μg ceftazidime disks (Oxoid Limited, Basingstoke, United Kingdom) on Mueller-Hinton agar with or without cloxacillin (Sigma-Aldrich, Taufkirchen, Germany) at 500 mg/liter as described elsewhere (8). The test was considered positive when the diameter of the inhibition zone around the ceftazidime disk increased by at least 10 mm in the presence of cloxacillin.
Competitive growth assays with mutant and parent strains.
The fitness of the mutants selected during the in vitro simulations in comparison to that of the parent strains was investigated. Selected mutants (retrieved from the last sample from the respective in vitro model) and the corresponding parent strain, each inoculated at 102 CFU/ml, were incubated at 37°C for 8 h in a batch culture in the absence of the antimicrobial agent. The P. aeruginosa mutants selected during the in vitro model experiments with PAO1 and meropenem, PAO1 and ceftazidime, and 12-09-15 and meropenem were designated PAO1/MEM, PAO1/CAZ, and 12-09-15/MEM, respectively.
PCR and sequencing experiments.
For P. aeruginosa 12-09-15, the mismatch repair system genes mutS, mutL, and uvrD were amplified and sequenced. Therefore, the following primers were used: mutS-f (5′-CTTCCGAAGGCCCGTATGA-3′), mutS-r (5′-TTGTGCGGTAGTCCGTCAGA-3′), mutS-s1 (5′-ATGGGACTTCGATCGCGA-3′), mutS-s2 (5′-ATCGGCACCTATCCCGAA-3′), mutS-s3 (5′-ACGACCTGGCGCTGGATGC-3′), mutL-f (5′-ATGAGTGAAGCACCGCGTAT-3′), mutL-r (5′-CGCAGGAAGAGCTTGTCCA-3′), mutL-s1 (5′-TGCACGAGGCGCGAGACGAGC-3′), mutlL-s2 (5′-TATACCCGGCCGGAGGCG-3′), uvrD-f (5′-ATGAACGACGACCTCTCCCTC-3′), uvrD-r (5′-CTACAGGGCTTCCAGCTTG-3′), uvrD-s1 (5′-ACCATCCCGGCGTGCTCGAGC-3′), and uvrD-s2 (5′-AACGACGCGGCGCTGGAACG-3′). Primers D1 (5′-CCTCAACAAGAGTGACCAAC-3′) and D2 (5′-TTACAGGATCGACAGCGGA-3′) were used to amplify and sequence the oprD genes from the parent strains P. aeruginosa PAO1 and P. aeruginosa 12-09-15 and the mutants P. aeruginosa PAO1/MEM and P. aeruginosa 12-09-15/MEM selected in the in vitro model experiments. For the amplification and sequencing of ampD from P. aeruginosa PAO1 and P. aeruginosa PAO1/CAZ, the previously described primers DE1 and DE2 were used (2). Sequencing was performed by SEQLAB Sequence Laboratories Göttingen GmbH, Göttingen, Germany.
RESULTS
Antibacterial effect.
The pharmacodynamic parameters and pharmacological indices and the killing kinetics for the in vitro model experiments conducted are presented in Table 1 and Fig. 1.
TABLE 1.
Pharmacodynamic parameters and pharmacokinetic/pharmacodynamic indicesa
| Antimicrobial agent | P. aeruginosa strain | MIC (mg/liter) | Rmax (Δlog10 CFU/ml) | Time (h) between initial dose and Rmax | AAC0-24 (Δlog10 CFU/ml·h) | T>MIC (%) | Cmax/MIC |
|---|---|---|---|---|---|---|---|
| Meropenem | PAO1 | 0.5 | −1.09 | 1 | −22.46 | 66.67 | 112.2 |
| 12-09-15 | 2 | −1.23 | 3 | −19.58 | 41.67 | 28.05 | |
| Ceftazidime | PAO1 | 2 | −1.28 | 17.5 | 9.29 | 100 | 86.9 |
| 12-09-15 | 16 | 45.75 | 10.86 |
Rmax, maximal bactericidal effect; AAC, area above the curve; T>MIC, cumulative percentage of time that the drug concentration exceeds the MIC; Cmax/MIC, peak level divided by the MIC; Δlog10 CFU/ml is the difference between the log10 CFU/ml at a given time and the log10 CFU/ml of the initial inoculum.
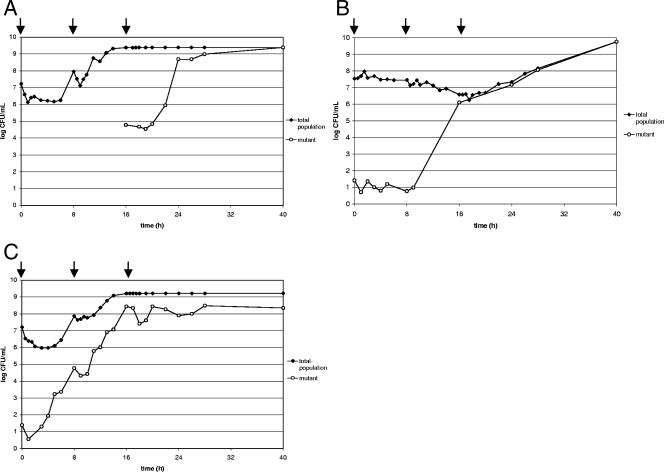
FIG. 1.
(A and B) CFU counts indicating the total populations of PAO1 cells and the populations of mutant cells during treatment with meropenem (three 1-g doses) (A) and during treatment with ceftazidime (three 2-g doses) (B). (C) CFU counts indicating the total population of 12-09-15 cells and the population of mutant cells during treatment with meropenem (three 1-g doses). Arrows indicate dosing times.
Antibacterial effect of meropenem.
The MIC of meropenem for P. aeruginosa PAO1 was 0.5 mg/liter. For P. aeruginosa 12-09-15, the meropenem MIC was two dilution steps higher (2 mg/liter), but the strain was still clinically susceptible according to CLSI breakpoints. The results of oprD sequencing and efflux pump inhibition testing for P. aeruginosa 12-09-15 revealed that the elevated meropenem MIC for this strain was not due to the loss of OprD but was determined by the overexpression of efflux pumps (Table 2). The Rmax for P. aeruginosa PAO1, achieved at 1 h after the first dose of 1 g of meropenem, was −1.09 Δlog10 CFU/ml, and the Rmax for P. aeruginosa 12-09-15, reached at 3 h, was −1.23 Δlog10 CFU/ml. The drug concentration fell below the MICs for P. aeruginosa PAO1 and P. aeruginosa 12-09-15 after 5.33 and 2.33 h, respectively. For both strains, bacterial regrowth occurred shortly afterwards, and both strains reached a cell count above the inoculum after 8 h. For P. aeruginosa 12-09-15, the second and third doses of meropenem showed hardly any effect at all. P. aeruginosa PAO1 showed a reduction of −0.83 Δlog10 CFU/ml after the second dose, but regrowth occurred as soon as that of P. aeruginosa 12-09-15 and was also not affected by the third dose. At 40 h after the first dose, the bacterial cell count was 20 times that of the inoculum for both strains.
TABLE 2.
Phenotypic and genotypic characterization of parent and mutant strains selected during treatment with meropenem and ceftazidimea
| Antimicrobial agent | P. aeruginosa strain | MIC (mg/liter) of:
|
Efflux pump inhibition test
|
Cephalosporinase inhibition test
|
Modification(s) in:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inhibition zone diam (mm) for CAZ (30 μg)
|
Difference in inhibition zone diam (mm) | Result | ||||||||||||||
| MEM | IMP | CAZ | PIP | PIP-TAZ | MIC (mg/liter) of LEV
|
Reduction of MIC (n-fold) | Result | Without cloxacillin | With cloxacillinc | OprD | AmpD | |||||
| Without Phe-Arg-β-naphthylamide | With Phe-Arg-β-naphthylamideb | |||||||||||||||
| MEM | PAO1 | 0.5 | 2 | 0.5 | 0.06 | 8 | − | None | ||||||||
| 12-09-15 | 2 | 4 | 128 | 2 | 64 | + | None | |||||||||
| PAO1/MEM | 4 | 16 | 0.5 | 0.12 | 4 | − | Insertion of C after A1205; frameshift | |||||||||
| 12-09-15/MEM | 16 | 16 | 64 | 1 | 64 | + | None | |||||||||
| CAZ | PAO1 | 2 | 4 | 4 | 0.5 | 0.06 | 8 | − | 27 | 27 | 0 | − | None | |||
| PAO1/CAZ | 32 | 64 | 64 | 0.5 | 0.06 | 8 | − | 17 | 27 | 10 | + | None | ||||
MEM, meropenem; IMP, imipenem; CAZ, ceftazidime; PIP, piperacillin; PIP-TAZ, piperacillin-tazobactam; LEV, levofloxacin; +, overproduction of efflux pumps or AmpC β-lactamase; −, no overproduction of efflux pumps or AmpC β-lactamase.
Phe-Arg-β-naphthylamide was included at 20 mg/liter.
Cloxacillin was included at 500 mg/liter.
Antibacterial effect of ceftazidime.
The MIC of ceftazidime for P. aeruginosa PAO1 was 2 mg/liter. The first application of 2 g of ceftazidime did not result in a reduction of the P. aeruginosa PAO1 cell count. After the second dose of ceftazidime, the cell count was slowly reduced by 1 order of magnitude, and the Rmax of −1.28 Δlog10 CFU/ml was detected after the third dose, at 17.5 h. Up to that time, slow bacterial regrowth occurred. During the treatment with ceftazidime, the drug concentration did not fall below the MIC (2 mg/liter) for the bacterial test strain P. aeruginosa PAO1 for 24 h. At 40 h after the first application of the drug, the P. aeruginosa PAO1 cell count was 20 times that of the inoculum. Because of unfavorable pharmacodynamic parameters and pharmacokinetic-pharmacodynamic indices for ceftazidime and P. aeruginosa 12-09-15 (MIC, 16 mg/liter; T>MIC, 45.75%), no in vitro simulation with this combination was carried out.
Selection of mutants.
The selection of mutants during the in vitro model experiments is depicted in Fig. 1.
Mutants of P. aeruginosa PAO1 selected during treatment with meropenem.
After the first application of meropenem, no mutant selection was detectable, as the count of mutant cells was below the detection limit. During the administration of the second dose, detectable mutant selection took place, resulting in a mutant population that constituted 0.026‰ of the total after 16 h. Following the third application of meropenem, the proportion of mutants increased rapidly, and at the end of the in vitro model simulation, only mutants were found in the population.
Mutants of P. aeruginosa PAO1 selected during treatment with ceftazidime.
Under the selective pressure of ceftazidime, mutant selection during the application of the second dose was also detected but to a 10,000-fold-greater extent than that during the application of the second dose of meropenem. After 16 h, at the beginning of the administration of the third dose, 33% of cells in the total population were mutant cells. Subsequently, the proportion of the detected mutants further increased, reaching 100% of the total population after 40 h.
Mutants of P. aeruginosa 12-09-15 selected during treatment with meropenem.
For P. aeruginosa 12-09-15, the selection of mutants started during the application of the first dose and yielded a mutant population of 0.811‰ of the total population after 8 h. The proportion of mutant cells further increased after the second application, to 16.7%. No further increase of the mutant cell numbers was observed from the administration of the last dose to the end of the in vitro model experiment at 40 h.
Characterization of hypermutability.
The mutation frequency for rifampin resistance in P. aeruginosa 12-09-15 was 2.26 ± 0.23 mutants/106 cells. The presence of mutations responsible for the hypermutation phenotype of P. aeruginosa 12-09-15 was investigated by sequencing the mismatch repair system genes mutS, mutL, and uvrD. mutL had a 1-bp deletion (A1250) in codon 417 that resulted in a frameshift. No mutations in mutS and uvrD were detected.
Competitive growth assays with mutant and parent strains.
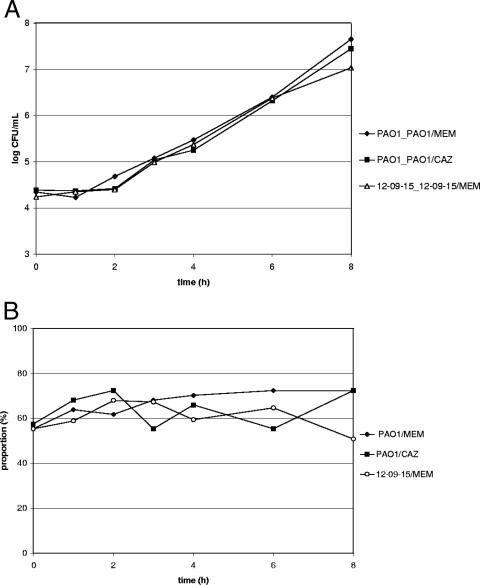
The mutants selected during the in vitro simulations, P. aeruginosa PAO1/MEM, P. aeruginosa PAO1/CAZ, and P. aeruginosa 12-09-15/MEM, showed no disadvantage in fitness in the absence of selective pressure. As depicted in Fig. 2, no significant changes in the proportions of mutants in the total population were observed during the course of the competitive growth assays.
FIG. 2.
Results of the competitive growth assays with parent and mutant strains selected in in vitro model experiments with P. aeruginosa PAO1 and meropenem (PAO1_PAO1/MEM), P. aeruginosa PAO1 and ceftazidime ((PAO1_PAO1/CAZ), and P. aeruginosa 12-09-15 and meropenem (12-09-15_12-09-15/MEM). CFU counts indicating the total populations (A) and the proportion of the respective mutant (B) are shown.
Phenotypic and genotypic characterization of selected mutants and parent strains.
The test strains P. aeruginosa PAO1 and P. aeruginosa 12-09-15 used for the meropenem in vitro models and their respective mutants selected during those experiments, P. aeruginosa PAO1/MEM and P. aeruginosa 12-09-15/MEM, were tested for carbapenem susceptibility, the overproduction of efflux pumps, and modifications in oprD.
For P. aeruginosa PAO1/MEM, the meropenem MIC was three dilution steps higher than that for the parent strain (4 mg/liter versus 0.5 mg/liter), but this mutant was still susceptible according to CLSI breakpoints. The decrease in the susceptibility of the mutant to imipenem resulted in resistance (MIC for PAO1, 2 mg/liter; MIC for PAO1/MEM, 16 mg/liter).
Contrary to P. aeruginosa PAO1/MEM, P. aeruginosa 12-09-15/MEM did not retain the susceptibility of the parent strain to meropenem but exhibited resistance (MIC for PAO1, 2 mg/liter; MIC for 12-09-15/MEM, 16 mg/liter), while the MIC of imipenem for this mutant also increased to the level of resistance (from 4 to 16 mg/liter). For P. aeruginosa PAO1 and P. aeruginosa PAO1/MEM, no overproduction of efflux pumps was proven. For the mutator strain P. aeruginosa 12-09-15, the test for efflux pump overproduction was positive, with a 64-fold reduction of the MIC of levofloxacin in the presence of the efflux pump inhibitor Phe-Arg-β-naphthylamide. The reduction of the MIC of levofloxacin for P. aeruginosa 12-09-15/MEM in the presence of the efflux pump inhibitor was equivalent to the reduction of the MIC for the parent strain (64-fold); therefore, no additional efflux pump overproduction occurred. The eight- and fourfold increases in the MICs of meropenem and imipenem, respectively, for PAO1/MEM and 12-09-15/MEM pointed to the loss of the porin OprD in both strains. For P. aeruginosa PAO1/MEM, sequencing revealed the loss of OprD due to a frameshift mutation in oprD. No mutation was detected in oprD of P. aeruginosa 12-09-15/MEM. However, an analysis of the outer membrane proteins of the parent strain, P. aeruginosa 12-09-15, and the mutant strain by sodium dodecyl sulfate-polyacrylamide gel electrophoresis clearly indicated that 12-09-15/MEM did not produce OprD or produced only very small amounts (data not shown).
For the test strain P. aeruginosa PAO1 and the mutant P. aeruginosa PAO1/CAZ used for and retrieved during the in vitro model experiment with ceftazidime, respectively, susceptibility to ceftazidime, piperacillin, and piperacillin-tazobactam; the overproduction of efflux pumps; the overproduction of the AmpC β-lactamase; and mutations in ampD were investigated. The MIC of ceftazidime for the mutant P. aeruginosa PAO1/CAZ selected under simulated clinical ceftazidime profiles showed an increase from 2 to 32 mg/liter in comparison to the MIC for the parent strain, resulting in resistance. This change was in concordance with the result of the cephalosporinase inhibition test, which proved the overproduction of AmpC β-lactamase by this mutant. Sequencing revealed no mutations in ampD of P. aeruginosa PAO1/CAZ, pointing to another underlying mechanism of AmpC overproduction. The efflux pump inhibition test revealed no selection of efflux pump overproduction during the treatment for P. aeruginosa PAO1/CAZ.
DISCUSSION
Our study was conducted to investigate the selection of mutants of wild-type and hypermutable P. aeruginosa strains under clinical pharmacokinetic profiles of meropenem and ceftazidime.
Considering only pharmacodynamic parameters like AACs, the treatment of the wild-type P. aeruginosa strain PAO1 with ceftazidime seemed to yield a better outcome than treatment with meropenem (9.29 Δlog10 CFU/ml·h instead of −22.46 Δlog10 CFU/ml·h). However, the first dose of meropenem yielded a pronouncedly higher level of bactericidal efficacy (AAC from 0 to 8 h [AAC0-8], 5.3 Δlog10 CFU/ml·h) than the first dose of ceftazidime (AAC0-8, −0.19 Δlog10 CFU/ml·h). The MIC of meropenem for the hypermutable P. aeruginosa strain 12-09-15 was fourfold higher than that for the wild-type strain PAO1 (2 mg/liter versus 0.5 mg/liter). Of the pharmacokinetic/pharmacodynamic indices, the Cmax/MIC ratio has been postulated to be a good predictor of therapeutic efficacy (33) for antibiotics that show concentration-dependent killing. As β-lactam antibiotics are supposed to kill in a time-dependent manner, the elevated meropenem MIC for P. aeruginosa strain 12-09-15 should not impair the antimicrobial effect. Accordingly, no significant difference in the outcome of meropenem treatment of the 12-09-15 and PAO1 strains was found (AACs, −19.58 Δlog10 CFU/ml·h versus −22.46 Δlog10 CFU/ml·h).
Besides the antibacterial effect on a bacterial strain, the extent of mutant selection plays an important role in ranking antimicrobial agents and dosing regimens. In a previous study, decreased susceptibility to meropenem and ceftazidime due to enrichment with resistant mutants during exposure to constant antimicrobial concentrations was detected only with hypermutable but not wild-type P. aeruginosa strains (30). In contrast, in our study mutants of P. aeruginosa PAO1 were detected after exposure to drugs used according to clinical pharmacokinetic profiles. This difference can be explained by the selective pressure associated with the simulated concentration-time curves for the antibiotics. Furthermore, we used a larger initial population (∼3 × 109 cells versus 2 × 104 to 4 × 104 cells) that was sufficient to harbor already several mutants that further accumulated during treatment. This higher cell count was comparable to conditions in the clinical setting, as cell counts of 108 to 1010 CFU/ml of sputum from cystic fibrosis patients with chronic P. aeruginosa infections are found (13).
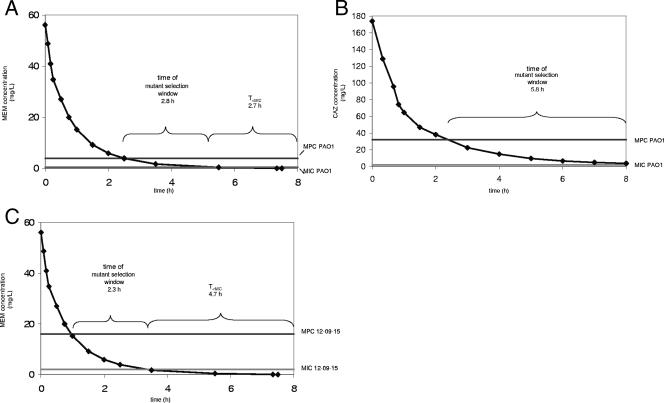
The ceftazidime-resistant one-step mutant P. aeruginosa PAO1/CAZ overproduced the AmpC β-lactamase due to an undetermined mechanism. AmpC overproduction without ascertained mechanisms is also found in clinical isolates (4), and more studies are necessary to elucidate the regulatory network involving ampC expression in P. aeruginosa. For P. aeruginosa PAO1/CAZ, the specific mutant selection window (lower boundary, MIC for the parent strain; upper boundary, MIC for the specific mutant as a surrogate parameter representing the MPC) lasted 5.8 h during each dosing interval (Fig. 3). The concentration of ceftazidime was not below the MIC for the parent strain P. aeruginosa PAO1 at any point during the whole dosing regimen. The competitive growth assay revealed no disadvantage for strain PAO1/CAZ in the absence of selective pressure. Therefore, the one-step mutant that was selected during each of the three mutant selection windows had no disadvantage compared to its parent strain at any time but continuously replaced the wild-type cells in the population.
FIG. 3.
The concentration-time curve (concentrations were taken from references 17 and 20), the duration of the mutant selection window, and the amount of time that the drug concentration was below the MIC (T<MIC) for the parent strain in each dosing interval for PAO1 and meropenem (MEM) (A), PAO1 and ceftazidime (CAZ) (B), and 12-09-15 and meropenem (C) are shown.
For P. aeruginosa PAO1/MEM, the duration of the specific mutant selection window (2.8 h) and the amount of time below the MIC (2.7 h) were approximately the same. P. aeruginosa PAO1/MEM also had no disadvantage in growth in comparison to the wild type. Therefore, for comparable reasons, this one-step mutant also had the opportunity to finally replace its parent strain. However, this mutant, which lost the porin OprD, showed only an intermediate but not a full resistance level to the drug by which it had been selected.
The treatment of P. aeruginosa 12-09-15 with meropenem enriched the population with the mutant P. aeruginosa 12-09-15/MEM only during three specific mutant selection windows lasting 2.3 h each. During each dosing interval, the concentration of meropenem was below the MIC for the parent strain approximately twice as long (4.7 h) as the duration of the mutant selection window, giving the parent strain the opportunity for successful growth. Its growth actually seemed to avoid further enrichment with the one-step mutants. According to Drlica (9), this finding contradicts the traditional teaching that mutant enrichment occurs at concentrations below the MIC. It is worth noting that, despite the hypermutator phenotype of P. aeruginosa 12-09-15, the selective pressure of meropenem enriched the cell population with a one-step mutant that acquired only one further mechanism of resistance to carbapenems, the deficiency in OprD. In contrast to PAO1/MEM, 12-09-15/MEM developed full meropenem resistance through the deficiency in OprD, as according to CLSI breakpoints, the clinically susceptible parent strain already showed decreased meropenem susceptibility due to upregulated efflux. The mutant did not further upregulate efflux pumps conferring additional cross-resistance, although meropenem is a substrate of several efflux systems. For the estimation of the expected number of mutants in a population, the accumulation of existing mutants and the production of new ones, which depends on the mutation rate, should be taken into account, as noted by Luria and Delbruck (21). Therefore, regarding the number of cells of the one-step mutant P. aeruginosa 12-09-15/MEM and the growth of this mutant during the simulated treatment, we cannot preclude that two-step mutants of the hypermutable parent strain were selected but might have remained undetected because of low cell counts. The in vivo emergence of a two-step mutant during ciprofloxacin treatment in a mouse model of lung infection with hypermutable P. aeruginosa was shown previously (24).
An optimal antibiotic dosing regimen shows a high level of antibacterial activity and low potential for stimulating resistance development. Our results indicate that the commonly used dosing regimens for meropenem and ceftazidime cannot avoid the selection of mutants of wild-type and hypermutable P. aeruginosa strains. Maintaining antibiotic concentrations above the MIC for the one-step mutant (time above the MPC) for a longer period than in present regimens might be beneficial. As hypermutability is very common in P. aeruginosa strains from cystic fibrosis patients and resistance is acquired predominantly through the accumulation of chromosomal mutations, the selection of mutants is especially disadvantageous for the repetitive treatment of these chronically infected patients.
Acknowledgments
We are grateful to Robert E. W. Hancock for providing us with P. aeruginosa PAO1. We thank Christine Fuhst and Antina Barger for their invaluable assistance with in vitro model techniques. We also thank Sonja Burak for her excellent technical assistance with outer membrane preparations.
B. Henrichfreise received a fellowship from the German national academic foundation (Studienstiftung des deutschen Volkes). Financial support was also obtained from AstraZeneca.
Footnotes
Published ahead of print on 6 August 2007.
REFERENCES
- 1.Alou, L., L. Aguilar, D. Sevillano, M. J. Gimenez, O. Echeverria, M. L. Gomez-Lus, and J. Prieto. 2005. Is there a pharmacodynamic need for the use of continuous versus intermittent infusion with ceftazidime against Pseudomonas aeruginosa? An in vitro pharmacodynamic model. J. Antimicrob. Chemother. 55:209-213. [DOI] [PubMed] [Google Scholar]
- 2.Bagge, N., O. Ciofu, M. Hentzer, J. I. Campbell, M. Givskov, and N. Hoiby. 2002. Constitutive high expression of chromosomal β-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 46:3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowker, K. E., H. A. Holt, R. J. Lewis, D. S. Reeves, and A. P. MacGowan. 1998. Comparative pharmacodynamics of meropenem using an in-vitro model to simulate once, twice and three times daily dosing in humans. J. Antimicrob. Chemother. 42:461-467. [DOI] [PubMed] [Google Scholar]
- 4.Bratu, S., D. Landman, J. Gupta, and J. Quale. 2007. Role of AmpD, OprF and penicillin-binding proteins in β-lactam resistance in clinical isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 56:809-814. [DOI] [PubMed] [Google Scholar]
- 5.Carmeli, Y., N. Troillet, G. M. Eliopoulos, and M. H. Samore. 1999. Emergence of antibiotic-resistant Pseudomonas aeruginosa: comparison of risks associated with different antipseudomonal agents. Antimicrob. Agents Chemother. 43:1379-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Document M7-A7. CLSI, Wayne, PA.
- 7.Cunha, B. A. 2002. Pseudomonas aeruginosa: resistance and therapy. Semin. Respir. Infect. 17:231-239. [DOI] [PubMed] [Google Scholar]
- 8.De Champs, C., L. Poirel, R. Bonnet, D. Siro, C. Chanal, J. Sirot, and P. Nordmann. 2002. Prospective survey of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolates in a French Hospital in 2000. Anitmicrob. Agents Chemother. 46:3031-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drlica, K. 2003. The mutant selection window and antimicrobial resistance. J. Antimicrob. Chemother. 52:11-17. [DOI] [PubMed] [Google Scholar]
- 10.Gibson, R. L., J. L. Burns, and B. W. Ramsey. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918-951. [DOI] [PubMed] [Google Scholar]
- 11.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasso, S. G., G. Meinardi, I. de Caneri, and V. Tarnassia. 1978. New in vitro model to study the effect of antibiotic concentration and rate of elimination of antimicrobial activity. Antimicrob. Agents Chemother. 13:570-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoiby, N., H. Krogh Johansen, C. Moser, Z. Song, O. Ciofu, and A. Kharazmi. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 3:23-35. [DOI] [PubMed] [Google Scholar]
- 14.Juan, C., M. D. Macia, O. Gutierrez, C. Vidal, J. L. Perez, and A. Oliver. 2005. Molecular mechanisms of β-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob. Agents Chemother. 49:4733-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juan, C., B. Moya, J. L. Perez, and A. Oliver. 2006. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level β-lactam resistance involves three AmpD homologues. Antimicrob. Agents Chemother. 50:1780-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keil, S., and B. Wiedemann. 1995. Mathematical corrections for bacterial loss in pharmacodynamic in vitro dilution models. Antimicrob. Agents Chemother. 39:1054-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krueger, W. A., J. Bulitta, M. Kinzig-Schippers, C. Landersdorfer, U. Holzgrabe, K. G. Naber, G. L. Drusano, and F. Soergel. 2005. Evaluation by Monte Carlo simulation of the pharmacokinetics of two doses of meropenem administered intermittently or as a continuous infusion in healthy volunteers. Antimicrob. Agents Chemother. 49:1881-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 19.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luethy, R., J. Blaser, A. Bonetti, H. Simmen, R. Wise, and W. Siegenthaler. 1981. Comparative multiple-dose pharmacokinetics of cefotaxime, moxalactam, and ceftazidime. Antimicrob. Agents Chemother. 20:567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luria, S., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macia, M. D., D. Blanquer, B. Togores, J. Sauleda, J. L. Perez, and A. Oliver. 2005. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob. Agents Chemother. 49:3382-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macia, M. D., N. Borrell, M. Segura, C. Gomez, J. L. Perez, and A. Oliver. 2006. Efficacy and potential for resistance selection of antipseudomonal treatments in a mouse model of lung infection by hypermutable Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:975-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mouton, J. W., and J. G. den Hollander. 1994. Killing of Pseudomonas aeruginosa during continuous and intermittent infusion of ceftazidime in an in vitro pharmacokinetic model. Antimicrob. Agents Chemother. 38:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ochs, M. M., M. P. McCusker, M. Bains, and R. E. Hancock. 1999. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob. Agents Chemother. 43:1085-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 29.Oliver, A., F. Baquero, and J. Blazquez. 2002. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43:1641-1650. [DOI] [PubMed] [Google Scholar]
- 30.Oliver, A., B. R. Levin, C. Juan, F. Baquero, and J. Blazquez. 2004. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob. Agents Chemother. 48:4226-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pai, H., J. W. Kim, J. Kim, J. H. Lee, K. W. Choe, and N. Gotoh. 2001. Carbapenem resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 45:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rustige, C., and B. Wiedemann. 1990. Antibacterial activity of lomefloxacin in a pharmacokinetic in vitro model. Antimicrob. Agents Chemother. 34:1107-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuck, E. L., and H. Derendorf. 2005. Pharmacokinetic/pharmacodynamic evaluation of anti-infective agents. Expert Rev. Anti-Infect. Ther. 3:361-373. [DOI] [PubMed] [Google Scholar]
- 34.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 35.Tam, V. H., A. N. Schilling, S. Neshat, K. Poole, D. A. Melnick, and E. A. Coyle. 2005. Optimization of meropenem minimum concentration/MIC ratio to suppress in vitro resistance of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:4920-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent, J. L. 2003. Nosocomial infections in adult intensive-care units. Lancet 361:2068-2077. [DOI] [PubMed] [Google Scholar]