Abstract
Structure-based design was used to develop a focused library of A-ring-modified diphenyl ether InhA inhibitors. From this library of analogs, two high-affinity alkyl-substituted diphenyl ethers, 6PP and 8PP, were selected for advanced study into their in vitro activity against Mycobacterium tuberculosis clinical isolates, their in vivo properties, and their signature response mode of action. 6PP and 8PP demonstrated enhanced activity against whole bacteria and showed activity in a rapid macrophage model of infection. In addition, transcriptional profiling revealed that the A-ring modifications of 6PP and 8PP increased the specificity of each analog for InhA. Both analogs had substantially longer half-lives in serum than did the parent compound, exhibited a fivefold reduction in cytotoxicity compared to the parent compound, and were well tolerated when administered orally at 300 mg/kg of body weight in animal models. Thus, the A-ring modifications increased the affinity and whole-cell specificity of the compounds for InhA and increased their bioavailability. The next step in optimization of the pharmacophore for preclinical evaluation is modification of the B ring to increase the bioavailability to that required for oral delivery.
Isoniazid (INH) is the single most effective chemotherapeutic for the treatment of tuberculosis. While the exquisite potency of INH may result from a complex mode of action that still remains to be delineated completely (15), it is known that INH affects cell wall biosynthesis via alterations of the mycobacterial type II fatty acid biosynthesis (FAS-II) pathway, one mechanism of which is through inhibition of InhA, the FAS-II enoyl reductase (1, 13, 15, 19). Although INH resistance is associated with mutations in InhA as well as KasA and the upstream regulatory region of the mabA-inhA operon (12, 15, 19), the vast majority of INH-resistant isolates contain mutations in the KatG catalase peroxidase protein responsible for the activation of INH (2, 9, 14, 15). Consequently, novel compounds with a distinct pharmacophore that inhibits InhA but does not require activation by KatG hold promise for the treatment of multidrug-resistant (MDR) clinical strains of Mycobacterium tuberculosis.
Previously, we reported the synthesis of compounds designed to explore the diphenyl ether pharmacophore as a potential antitubercular agent. These studies were based on the known ability of triclosan to inhibit the enoyl reductase class of enzymes (20). Using rational drug development strategies, this initial study substantiated the use of modeling, enzyme inhibition, and whole-cell assays to identify compounds with enhanced activity against clinical strains of M. tuberculosis with various drug resistance profiles. However, it did not address the whole bacterial mode of action, toxicity, or in vivo activity of the compounds. Accordingly, the work presented here expands on our previous report by investigation of the mode of action and potential detoxification transcriptional patterns of the most potent analogs identified and by analysis of their efficacy in models of infection. Our findings demonstrate that although these high-affinity InhA inhibitors have issues with low bioavailability, they are more effective in vitro inhibitors with less cytotoxicity than the parent compound triclosan, thus narrowing the spectrum of structural changes required for drug enhancement. The transcriptional responses confirm that the alkyl diphenyl ethers inhibit InhA within the cell, unlike triclosan, which likely has other targets (6). The identification of a transcriptional response specific to the inhibition of InhA will be critical for the development of the next generation of high-affinity InhA inhibitors with improved in vivo properties.
MATERIALS AND METHODS
MIC determinations and cytotoxicity testing.
MICs were determined using the microplate dilution method as previously described (16). African green monkey kidney cells (Vero cells) were grown in RPMI 1640 medium supplemented with 1.5 g/liter sodium bicarbonate, 10 ml/liter 100 mM sodium pyruvate, 140 ml/liter 100× nonessential amino acids, 100 ml/liter penicillin-streptomycin solution (10,000 IU/10,000 μg/ml), and 10% bovine calf serum at 37°C in a 5% CO2 incubator with 75% humidity. Testing was conducted for 72 h at 37°C in a 5% CO2 incubator. Cells were washed, CellTiter 96 AQueous One solution was added to each well, and plates were incubated for 4 h at 37°C. Plates were read at 490 nm using a spectrophotometric plate reader, and the absorbance readings were used to calculate the 50% lethal concentration (LC50).
Rapid macrophage assay.
A rapid macrophage assay was developed to assess the activities of compounds against intracellular bacteria. J774A.1 cells were allowed to phagocytose M. tuberculosis H37Rv cells (multiplicity of infection of 1:10) for 4 h at 37°C and then washed four times with phosphate-buffered saline (PBS) and culture growth medium without (control) or with one of the two most potent compounds (6PP and 8PP) at the MIC or twice the MIC (2× MIC). At time zero and 1 and 4 days postinfection, triplicate monolayers per compound were washed four times with PBS, lysed by the addition of 0.1% Triton X-100, and plated on 7H11 medium. Following incubation at 37°C, CFU were enumerated.
In vivo evaluation.
The oral bioavailability of the compounds was tested using a bioassay approach, as described before (8). Eight to 10-week-old female C57BL/6 mice were dosed via oral gavage. Twenty minutes, 1 h, 2 h, and 4 h after being dosed, three mice were bled from the tail vein. Sera and standards of the compounds were tested in threefold dilutions against M. tuberculosis H37Rv. Bacterial growth was determined by measuring the optical density after 3, 6, 9, and 12 days. Estimations of serum drug levels (in μg ml−1 serum) were obtained by using the MIC data from the standard drug lanes.
Transcriptional profiling.
M. tuberculosis H37Rv was treated with 15 μM 6PP, 12 μM 8PP, or 86 μM triclosan or left untreated at 37°C with shaking for 2 h. Whole bacteria were subjected to TRIzol extraction, and total RNA was isolated by physical disruption. Microarray analysis was performed with labeled cDNAs generated using direct labeling from 5 μg of total RNA as described previously (17). The resulting fluorescence for each channel of the array (Cy3 and Cy5) was normalized to the mean channel intensity and analyzed using Genesifter analysis software. t test statistical analysis and Benjamini and Hochberg correction were applied to all analyses of all mean normalized data. Significance was considered to be a >1.5-fold alteration in expression, with a P value cutoff of <0.05. The transcriptional activity of selected genes identified by DNA microarray analysis was verified using real-time PCR as described previously (20).
RESULTS
Determination of 6PP and 8PP activities against clinical MDR strains.
We previously reported whole bacterial activity of a series of alkyl-disubstituted diphenyl ethers against the laboratory strain H37Rv and five clinical M. tuberculosis strains (Table 1) (20). The two most potent compounds, 6PP and 8PP, were nanomolar inhibitors of InhA and had MICs of 1 to 2 μg/ml against drug-sensitive and drug-resistant M. tuberculosis. In general, gene dosage increased the MICs for all compounds; however, the MIC was influenced more for analogs substituted with short-chain alkyl groups than for triclosan or any of the long-chain-substituted compounds (Table 1). This observation indicates that there is a narrow range of alkyl substitutions that provide greater whole-cell specificity for InhA.
TABLE 1.
MICs and toxicity of alkyl diphenyl ethersa
| Compound | MIC99, μg ml−1 (μM)
|
LC50 (μg ml−1) | Therapeutic index | ||||||
|---|---|---|---|---|---|---|---|---|---|
| H37Rv | pMH29:InhA | W210 | TN587 | NHN20 | HN335-2 | NHN382 | |||
| Triclosan | 12.5 ± 0 (43.1 ± 0) | 33.3 ± 12.9 (115 ± 45) | 14.7 ± 3.8 (50.8 ± 13) | 12.5 ± 0 (43.1 ± 0) | 12.5 ± 0 (43.1 ± 0) | 18.8 ± 6.3 (64.9 ± 21.7) | 12.5 ± 0 (43.1 ± 0) | <12.5 | <1 |
| 2PP | 15.6 ± 7.7 (72.9 ± 35.7) | 133.3 ± 57.7 (622.3 ± 269.5) | 10.4 ± 3.2 (48.6 ± 15.1) | 8.9 ± 4.2 (41.3 ± 19.4) | 14.6 ± 8.5 (68.1 ± 39.9) | 6.3 ± 53.4 (629.2 ± 16) | 10.4 ± 3.2 (48.6 ± 15.1) | <15.6 | <1 |
| 4PP | 2.6 ± 0.8 (10.8 ± 3.3) | 50.0 ± 0 (206.4 ± 0) | 2.6 ± 0.8 (10.8 ± 3.3) | 2.1 ± 0.8 (8.7 ± 3.3) | 2.1 ± 0.9 (8.7 ± 3.3) | 2.8 ± 0.7 (11.6 ± 2.8) | 3.8 ± 1.4 (15.5 ± 5.8) | 1.6 | <1 |
| 5PP | 4.2 ± 1.6 (116.3 ± 6.3) | 50.0 ± 0 (195.1 ± 0) | 1.6 ± 0 (6.2 ± 0) | 1.6 ± 1 (6.2 ± 0) | 1.6 ± 2 (6.2 ± 0) | 2.1 ± 0.8 (8.2 ± 3.1) | 4.2 ± 1.6 (16.3 ± 6.3) | 3.8 | 1 |
| 6PP | 2.1 ± 0.9 (7.8 ± 3.3) | 18.8 ± 6.8 (69 ± 25) | 2.9 ± 0.4 (10.7 ± 1.5) | 2.0 ± 1.0 (7.4 ± 3.7) | 3.1 ± 0 (11.5 ± 0) | 3.7 ± 0.9 (13.7 ± 3.3) | 3.1 ± 0 (11.5 ± 0) | 12.5 | 6 |
| 8PP | 1.9 ± 0.5 (6.4 ± 1.7) | 22.9 ± 5.1 (77 ± 17) | 2.6 ± 0.4 (8.7 ± 1.3) | 2.0 ± 1.0 (6.7 ± 3.4) | 2.4 ± 0.76 (8.0 ± 2.6) | 3.1 ± 0 (10.4 ± 0) | 2.6 ± 0.9 (8.7 ± 3.0) | 9.9 | 5 |
| 9PP | 14.1 ± 9.2 (45 ± 29.5) | 50.0 ± 0 (160.0 ± 0) | 8.3 ± 3.2 (26.7 ± 10.3) | 7.3 ± 4.3 (23.3 ± 13.7) | 16.7 ± 6.5 (53.3 ± 20.7) | 12.5 ± 0 (40 ± 0) | 4.7 ± 1.8 (15 ± 5.8) | <14.1 | <1 |
The laboratory strain M. tuberculosis H37Rv and clinical strains W210 (KatG wild type), TN587 (KatG-S315T), and NHN382 (KatG-Del), with differing drug resistance profiles, were used for MIC determinations. Toxicity was evaluated using Vero cells, and the therapeutic index was calculated as LC50/MIC99. Data are means ± standard deviations.
Evaluation of cytotoxicity of diphenyl ether analogs.
The diphenyl ether analogs with MICs of <50 μg/ml were tested for general cytotoxicity using a Vero cell line (Table 1). Many of the compounds had LC50 values at their MIC concentration. However, 6PP and 8PP had LC50 values of 13 μg/ml and 10 μg/ml, resulting in therapeutic indexes of 6 and 5, respectively. In contrast, the parent compound had an LC50 at or below the MIC, resulting in a measurable therapeutic index of 1.
Assessment of serum binding and of activity in a rapid macrophage assay.
To evaluate whether protein binding alters the activities of 6PP and 8PP and whether they are able to inhibit the growth of intracellular M. tuberculosis, the MICs of these compounds were determined in the presence of 10% mouse serum (according to CLSI guidelines), and their activity against bacteria was evaluated in a rapid macrophage assay. The results showed that the MIC for 6PP or 8PP was not influenced by the presence of serum, indicating that neither of these compounds has serum binding problems (Table 2). Furthermore, the addition of 6PP or 8PP at the 2× MIC to macrophages containing M. tuberculosis resulted in bacterial growth inhibition of 73% ± 7% and 71% ± 1%, respectively, which is demonstrative of intracellular antimycobacterial activity. INH was included as a positive control and reduced growth by 98% ± 1% at the 2× MIC. The inhibitory activities of 6PP and 8PP against intracellular bacteria in the rapid macrophage model are similar to those observed for other compounds tested for activity against M. tuberculosis (18). It should also be noted that both 6PP and 8PP displayed much lower toxicity in the macrophage assay than did the parent compound, triclosan, which was lethal to macrophages after 1 day of treatment at both the MIC and 2× MIC levels.
TABLE 2.
Protein binding and bioavailability analysis of 6PP and 8PPa
| Compound | Dose (mg kg−1) | Formulation | Method of compound delivery | MIC (μg ml−1)
|
Drug serum level (μg ml−1) | Serum absorption index | |
|---|---|---|---|---|---|---|---|
| Without serum | With serum | ||||||
| 6PP | 300 | Methylcellulose | Oral | 3.3 | 3.3 | 66 | 20 |
| 6PP | 300 | Cyclodextrin | Oral | 3.3 | 3.3 | 132 | 40 |
| 6PP | 300 | PBS | Subcutaneous | 3.3 | 3.3 | 99 | 30 |
| 8PP | 300 | Methylcellulose | Oral | 1.1 | 1.1 | 20 | 18 |
| Triclosan | 300 | PBS | Subcutaneous | 3.3 | 10 | 66 | 30 |
| INH | 25 | H2O | Oral | 0.05 | 0.05 | >13 | >320 |
In vitro MICs for H37Rv were determined within the bioavailability assay by broth microdilution. Serum (10%) was added to one set of controls to determine the effect of protein binding on the MIC. Drug levels in mouse serum were estimated by multiplying the dilution factor by the MIC of the drug in the absence of serum. The serum absorption index was calculated as the serum concentration divided by the MIC99 and is a measure of the bioavailability of the compound.
Bioavailability and in vivo efficacy of 6PP and 8PP.
As a prelude to more detailed in vivo studies, a bioavailability assay was performed to assess the serum levels of 6PP and 8PP after oral administration (8). Drug serum levels of 6PP and 8PP ranged from 66 to 132 μg/ml depending on the formulation (Table 2), resulting in serum absorption indices of 18 to 40. This is in contrast to INH, which had serum levels of >13 μg/ml, resulting in serum absorption indices of >320. Importantly, the presence of analogs could be detected in the serum 8 h after dosing, which is much longer than the time to detection reported for INH (3).
Transcriptional differences between triclosan, 6PP, and 8PP.
Previous reports indicate that triclosan affects respiration in addition to fatty acid synthesis, which likely results because triclosan is only a modest InhA inhibitor and thus must be used at concentrations that elicit other cellular effects (6). In order to determine if the increased potencies of 6PP and 8PP towards InhA had narrowed the mode of action of these compounds, the global transcriptional response of H37Rv treated with 6PP, 8PP, or triclosan was assessed. These studies showed that triclosan had a more pleiotropic effect on bacterial metabolism than did treatment with either 6PP or 8PP (Table 3; see Tables S1 and S2 in the supplemental material). In particular, 6PP and 8PP upregulated hallmark genes associated with cell wall synthesis, including fas, the KAS operon, accD4, pks13, pks16, fadD32, and rv0241c (6), demonstrating that these compounds specifically target fatty acid biosynthesis. In contrast, triclosan failed to induce cell wall synthesis genes; rather, this compound induced a large number of genes involved in β-oxidation, including putative acyl-coenzyme A (acyl-CoA) synthase genes, genes for acyl-CoA dehydrogenases that catalyze the initiation of β-oxidation, and genes that encode β-oxidation proteins responsible for cyclic degradation of fatty acids. In addition, 6PP and 8PP also induced genes involved in meromycolate modification (umaA, mmaA3, and mmaA4), genes involved in the synthesis of arabinogalactan (embA and embB), the final cell wall acceptor for mycolic acids, and the gene for a member of the antigen 85 complex (fpbC) involved in deposition of mycolic acids (5).
TABLE 3.
Summary of differentially regulated ORFs, according to functional classification, after drug treatmenta
| Drug | Total no. of ORFs analyzed | Total no. of differentially regulated ORFsb | No. (%) of differentially regulated ORFs
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell wall and cell processes | Conserved hypothetical | Information pathways | Insertion sequences and phages | Intermediary metabolism and respiration | Lipid metabolism | PE/PPE | Regulatory proteins | Unknown | Virulence, detoxification, adaptation | |||
| Triclosan | 2,498 | 722 | 93 (17) | 145 (19) | 77 (10) | 15 (2) | 15 (25) | 62 (8) | 33 (4) | 25 (3) | 107 (14) | 20 (3) |
| 6PP | 737 | 127 | 21 (10) | 31 (24) | 4 (3) | 3 (2) | 27 (21) | 11 (9) | 3 (2) | 12 (9) | 7 (6) | 8 (6) |
| 8PP | 780 | 121 | 12 (10) | 38 (31) | 2 (2) | 4 (3) | 15 (12) | 14 (12) | 4 (3) | 13 (11) | 14 (12) | 5 (4) |
Functional classifications were those annotated by http://genolist.pasteur.fr/TubercuList/.
ORFs with expression differences of ≥1.5-fold (P > 0.05).
Previously, we reported that 6PP and 8PP failed to upregulate a putative efflux pump (rv1685c-rv1687c) and aromatic dioxygenase (rv3160c-rv3161c) that were induced by triclosan. The additional studies reported here now show that, instead, 6PP and 8PP induce the expression of iniABC, an operon reported to encode at least one component of an undefined efflux mechanism that is known to be induced by and associated with tolerance to the cell wall inhibitors INH and ethambutol (7). Thus, the diphenyl ether analogs circumvent detoxification mechanisms associated with triclosan but may induce a potential efflux pump associated with drug tolerance. Furthermore, treatment with 6PP and 8PP consistently caused increased transcription of rv2846c (efpA), encoding an efflux protein which leads to decreased susceptibility to INH when deleted in Mycobacterium smegmatis (11). This particular efflux protein was not upregulated in M. tuberculosis following treatment with the parent compound, triclosan. These studies again raise important differences between triclosan and the alkyl diphenyl ethers and also underline the similarity in transcriptional responses between the latter and cell wall biosynthesis inhibitors such as INH.
High-content multiple-feature profiles of high-affinity InhA inhibitors.
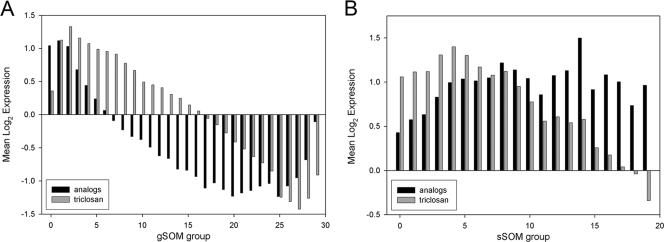
Tandem self-organizing map (tandem-SOM) analysis was performed to identify open reading frames (ORFs) that can be used to construct high-content multiple-feature screens to monitor gene responses of next-generation diphenyl ethers (Fig. 1). When the signature response of M. tuberculosis to 6PP and 8PP treatment was studied and deciphered, genes (n = 60) that were uniquely responsive to treatment with diphenyl ethers (≥1.5-fold change in expression) were grouped in global SOM (gSOM) groups 0 to 1 and genes that were induced by treatment with the analogs were grouped in sub-SOM (sSOM) groups 7 to 19. These genes are listed in Table 4 and include genes encoding fatty acid and mycolic acid synthases. This provides a signature profile for identifying novel compounds that have a similar mode of action to 6PP and 8PP.
FIG. 1.
Tandem-SOM analysis. (A) gSOM analysis of transcriptionally active ORFs. (B) sSOM analysis of ORFs from groups 0 to 2 of gSOM analysis. gSOM analysis distributed 3,627 ORFs into 30 groups (groups 0 to 29). sSOM analysis distributed 803 ORFs into 20 groups (groups 0 to 19).
TABLE 4.
High-content multiple-feature profiles of high-affinity InhA inhibitorsa
| ORF | Gene name | gSOM group | sSOM group | Ratio | P value | Protein name or function |
|---|---|---|---|---|---|---|
| Rv0129c | fbpC2 | 0 | 14 | 3.4 | 0.0049 | Secreted antigen 85 complex C (FbpC) |
| Rv0171 | Rv0171 | 0 | 15 | 1.5 | 0.0476 | Mce family protein Mce1C |
| Rv0172 | Rv0172 | 0 | 13 | 1.5 | 0.0217 | Mce family protein Mce1D |
| Rv0179c | lprO | 0 | 17 | 1.8 | 0.0070 | Possible lipoprotein LprO |
| Rv0180c | Rv0180c | 0 | 9 | 1.7 | 0.0107 | Probable conserved transmembrane protein |
| Rv0181c | Rv0181c | 1 | 8 | 1.7 | 0.0072 | Hypothetical protein |
| Rv0197 | Rv0197 | 0 | 13 | 1.7 | 0.0029 | Possible oxidoreductase |
| Rv0342 | iniA | 0 | 15 | 1.5 | 0.0050 | INH-inducible gene protein IniA |
| Rv0410c | pknG | 0 | 17 | 1.5 | 0.0499 | Serine/threonine protein kinase G (STPK G) |
| Rv0411c | glnH | 0 | 16 | 1.8 | 0.0040 | Probable glutamine-binding lipoprotein GlnH (GlnBP) |
| Rv0412c | Rv0412c | 0 | 16 | 1.6 | 0.0044 | Possible conserved membrane protein |
| Rv0474 | Rv0474 | 0 | 13 | 2.4 | 0.0001 | Probable transcriptional regulatory protein |
| Rv0676c | mmpL5 | 0 | 9 | 2.4 | 0.0043 | Probable conserved transmembrane transport protein MmpL5 |
| Rv0678 | Rv0678 | 1 | 7 | 2.9 | 0.0163 | Hypothetical protein |
| Rv0680c | Rv0680c | 0 | 8 | 1.7 | 0.0225 | Probable conserved transmembrane protein |
| Rv0724 | sppA | 0 | 12 | 1.6 | 0.0007 | Possible protease IV SppA (endopeptidase IV) |
| Rv0805 | Rv0805 | 0 | 13 | 1.5 | 0.0050 | Hypothetical protein |
| Rv0846c | Rv0846c | 0 | 14 | 1.7 | 0.0046 | Probable oxidase |
| Rv0896 | gltA2 | 0 | 10 | 1.5 | 0.0246 | Citrate synthase |
| Rv0932c | pstS | 0 | 14 | 1.6 | 0.0172 | Periplasmic phosphate-binding lipoprotein PstS2 (PBP-2) |
| Rv1022 | lpqU | 1 | 7 | 2.3 | 0.0256 | Probable conserved lipoprotein LpqU |
| Rv1130 | Rv1130 | 0 | 17 | 3.9 | 0.0117 | Hypothetical protein |
| Rv1131 | Rv1131 | 0 | 19 | 2.5 | 0.0001 | Citrate synthase |
| Rv1157c | Rv1157c | 0 | 16 | 2.0 | 0.0037 | Conserved hypothetical Ala-, Pro-rich protein |
| Rv1158c | Rv1158c | 0 | 16 | 2.2 | 0.0116 | Conserved hypothetical Ala-, Pro-rich protein |
| Rv1218c | Rv1218c | 0 | 12 | 1.6 | 0.0001 | ATP-binding protein ABC transporter |
| Rv1286 | cysN | 0 | 9 | 2.5 | 0.0341 | Bifunctional sulfate adenylyltransferase |
| Rv1292 | argS | 0 | 10 | 1.5 | 0.0326 | Arginyl-tRNA synthetase |
| Rv1397c | Rv1397c | 0 | 16 | 1.5 | 0.0045 | Hypothetical protein |
| Rv1416 | ribH | 1 | 7 | 1.8 | 0.0251 | Riboflavin synthase subunit beta |
| Rv1497 | lipL | 0 | 13 | 1.7 | 0.0009 | Probable esterase LipL |
| Rv1548c | PPE | 0 | 12 | 1.5 | 0.0413 | PPE family protein |
| Rv1690 | lprJ | 0 | 11 | 2.1 | 0.0008 | Probable lipoprotein LprJ |
| Rv1702c | Rv1702c | 1 | 7 | 1.7 | 0.0266 | Hypothetical protein |
| Rv1782 | Rv1782 | 0 | 13 | 1.5 | 0.0010 | Probable conserved membrane protein |
| Rv1784 | Rv1784 | 0 | 12 | 1.6 | 0.0007 | Hypothetical protein |
| Rv1875 | Rv1875 | 0 | 11 | 1.5 | 0.0167 | Hypothetical protein |
| Rv1880c | Rv1880c | 1 | 7 | 1.6 | 0.0344 | Probable cytochrome P450 140 (CYP140) |
| Rv1912c | fadB5 | 1 | 8 | 3.0 | 0.0319 | Possible oxidoreductase FadB5 |
| Rv1945 | Rv1945 | 0 | 10 | 1.8 | 0.0087 | Hypothetical protein |
| Rv2053c | Rv2053c | 0 | 10 | 2.1 | 0.0030 | Probable transmembrane protein |
| Rv2193 | ctaE | 0 | 10 | 1.5 | 0.0445 | Probable cytochrome c oxidase (subunit III) (CtaE) |
| Rv2234 | ptpA | 1 | 8 | 1.5 | 0.0144 | Phosphotyrosine protein phosphatase PtpA |
| Rv2454c | Rv2454c | 0 | 12 | 1.5 | 0.0213 | Ferrodoxin oxidoreductase beta subunit |
| Rv2516c | Rv2516c | 0 | 8 | 2.2 | 0.0388 | Hypothetical protein |
| Rv2963 | Rv2963 | 0 | 15 | 1.7 | 0.0025 | Probable integral membrane protein |
| Rv3140 | fadE23 | 0 | 12 | 2.1 | 0.0005 | Probable acyl-CoA dehydrogenase FadE23 |
| Rv3159c | PPE | 1 | 8 | 2.0 | 0.0367 | PPE family protein |
| Rv3209 | Rv3209 | 0 | 14 | 1.8 | 0.0016 | Conserved hypothetical threonine- and proline-rich protein |
| Rv3249c | Rv3249c | 0 | 14 | 2.0 | 0.0055 | Transcriptional regulatory protein (probably TetR family) |
| Rv3251c | rubA | 0 | 14 | 2.0 | 0.0020 | Probable rubredoxin RubA |
| Rv3252c | Rv3252c | 0 | 13 | 2.1 | 0.0008 | Probable transmembrane alkane 1-monooxygenase AlkB |
| Rv3408 | Rv3408 | 0 | 13 | 1.8 | 0.0161 | Hypothetical protein |
| Rv3496c | Rv3496c | 0 | 13 | 1.5 | 0.0121 | Mce family protein Mce4D |
| Rv3500c | Rv3500c | 0 | 8 | 2.1 | 0.0427 | Conserved hypothetical integral membrane protein YrbE4B |
| Rv3740c | Rv3740c | 0 | 19 | 1.6 | 0.0013 | Hypothetical protein |
| Rv3741c | Rv3741c | 0 | 19 | 2.9 | 0.0002 | Possible oxidoreductase |
| Rv3810 | pirG | 0 | 12 | 2.2 | 0.0086 | Exported repetitive protein precursor PirG (EXP53) |
| Rv3837c | Rv3837c | 0 | 19 | 1.7 | 0.0117 | Probable phosphoglycerate mutase |
ORFs were identified by tandem-SOM analysis and analyzed statistically using Genesifter software. ORFs in bold were confirmed by quantitative real-time PCR with independent biological replicates. All ORFs listed were upregulated.
DISCUSSION
The development of novel drugs against M. tuberculosis has numerous hurdles; among them are identification of therapeutically relevant metabolic targets, optimization of potency, reduction of toxicity, and introduction of favorable physiochemical characteristics. Although the mode of action of INH is arguably complex, it is clear that InhA is a validated target for drug discovery (1, 4, 10, 12, 15, 19). Therefore, our strategy is to utilize this knowledge to develop new InhA inhibitors that are distinct in structure from INH and thus do not require activation by KatG. Through molecular modeling and target-based screening, a series of diphenyl ethers were synthesized, leading to the identification of two compounds as potential antimycobacterial drug candidates (20).
An area of concern regarding the development of novel inhibitors against an established target is that these compounds might also be ineffective against existing drug-resistant strains. However, by synthesizing high-affinity InhA inhibitors that did not require KatG activation, both 6PP and 8PP were fully active against representative MDR clinical isolates with various resistance profiles. In addition, given the necessary long duration of treatment, toxicity is a significant issue with all antimycobacterial drugs. However, the A-ring modifications led to a >5-fold reduction in cytotoxicity compared to that of triclosan.
Lower toxicity levels were also apparent upon treatment of macrophages, as triclosan was toxic to macrophages, while 6PP and 8PP were not. Importantly, however, both 6PP and 8PP showed the ability to reduce bacterial growth in the rapid macrophage assay, thus indicating that these compounds were able to enter the macrophage and remain effective against intracellular M. tuberculosis growing under altered metabolic conditions. Given orally, neither 6PP nor 8PP had bioavailability as high as that of INH, but the analogs could be detected at 8 h postadministration. In addition, there were no adverse reactions noted upon oral administration, substantiating the reduced toxicity of these compounds. Importantly, the increased serum half-lives of 6PP and 8PP compared to that of INH are expected to provide more constant drug pressure, allowing greater dosing intervals during treatment. However, because of the poor bioavailability of 6PP and 8PP, neither demonstrated significant efficacy in a rapid mouse model when delivered by gavage (data not shown). Thus, while not active orally, these compounds exhibit decreased cytotoxicity and increased half-lives, which are crucial characteristics for any newly developed drug. Together, the in vivo studies indicate that 6PP and 8PP have low cytotoxicity levels and can inhibit intracellular organisms but still have limited bioavailability.
The next step in optimizing the drug activity of diphenyl ethers is to combine mechanistic and chemical information so that physiochemical properties of diphenyl ethers can be modified for greater bioavailability without compromising their specificity. Previous studies have reported on the ability of signature transcriptional profiles to categorize drug classes based on metabolic pressure (6). The identification of an increased number of genes allows for a greater number of features to be interrogated, thus providing a higher statistical significance for signature profiling of a drug's mode of action. Thus, the next generation of compounds will be subjected to high-content multiple-feature profiling to prioritize screening efforts to those analogs with improved in vivo properties that maintain the desired metabolic effect on bacteria.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 AI-55298, R01 AI-044639, U01 AI-070383, NO1 AI-95385, and U54 AI-065357.
We acknowledge the postgenomic resources and services provided by the Rocky Mountain Regional Center of Excellence.
Footnotes
Published ahead of print on 30 July 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Banerjee, A., E. Dubnau, A. Quemard, V. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. D. Lisle, and W. R. Jacobs, Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230. [DOI] [PubMed] [Google Scholar]
- 2.Barry, C. E., III, R. E. Lee, K. Mdluli, A. E. Sampson, B. G. Schroeder, R. A. Slayden, and Y. Yuan. 1998. Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37:143-179. [DOI] [PubMed] [Google Scholar]
- 3.Barry, C. E., III, R. A. Slayden, A. E. Sampson, and R. E. Lee. 2000. Use of genomics and combinatorial chemistry in the development of new antimycobacterial drugs. Biochem. Pharmacol. 59:221-231. [DOI] [PubMed] [Google Scholar]
- 4.Basso, L. A., R. Zheng, J. M. Musser, W. R. Jacobs, Jr., and J. S. Blanchard. 1998. Mechanisms of isoniazid resistance in Mycobacterium tuberculosis: enzymatic characterization of enoyl reductase mutants identified in isoniazid-resistant clinical isolates. J. Infect. Dis. 178:769-775. [DOI] [PubMed] [Google Scholar]
- 5.Belisle, J. T., V. D. Vissa, T. Sievert, K. Takayama, P. J. Brennan, and G. S. Besra. 1997. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276:1420-1422. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff, H. I., T. G. Myers, B. R. Copp, M. R. McNeil, M. A. Wilson, and C. E. Barry III. 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J. Biol. Chem. 279:40174-40184. [DOI] [PubMed] [Google Scholar]
- 7.Colangeli, R., D. Helb, S. Sridharan, J. Sun, M. Varma-Basil, M. H. Hazbon, R. Harbacheuski, N. J. Megjugorac, W. R. Jacobs, Jr., A. Holzenburg, J. C. Sacchettini, and D. Alland. 2005. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol. Microbiol. 55:1829-1840. [DOI] [PubMed] [Google Scholar]
- 8.Gruppo, V., C. M. Johnson, K. S. Marietta, H. Scherman, E. E. Zink, D. C. Crick, L. B. Adams, I. M. Orme, and A. J. Lenaerts. 2006. Rapid microbiologic and pharmacologic evaluation of experimental compounds against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 50:1245-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heym, B., P. M. Alzari, N. Honore, and S. T. Cole. 1995. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol. 15:235-245. [DOI] [PubMed] [Google Scholar]
- 10.Kremer, L., J. D. Douglas, A. R. Baulard, C. Morehouse, M. R. Guy, D. Alland, L. G. Dover, J. H. Lakey, W. R. Jacobs, Jr., P. J. Brennan, D. E. Minnikin, and G. S. Besra. 2000. Thiolactomycin and related-analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. J. Biol. Chem. 275:16857-16864. [DOI] [PubMed] [Google Scholar]
- 11.Li, X. Z., L. Zhang, and H. Nikaido. 2004. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48:2415-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mdluli, K., R. A. Slayden, Y. Zhu, S. Ramaswamy, X. Pan, D. Mead, D. D. Crane, J. M. Musser, and C. E. Barry III. 1998. Inhibition of a Mycobacterium tuberculosis beta-ketoacyl ACP synthase by isoniazid. Science 280:1607-1610. [DOI] [PubMed] [Google Scholar]
- 13.Quemard, A., J. C. Sacchettini, A. Dessen, C. Vilcheze, R. Bittman, J. William, R. Jacobs, and J. S. Blanchard. 1995. Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry 34:8235-8241. [DOI] [PubMed] [Google Scholar]
- 14.Rouse, D. A., Z. Li, G.-H. Bai, and S. L. Morris. 1995. Characterization of the katG and inhA genes of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 39:2472-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slayden, R. A., and C. E. Barry III. 2000. The genetics and biochemistry of isoniazid resistance in Mycobacterium tuberculosis. Microbes Infect. 2:659-669. [DOI] [PubMed] [Google Scholar]
- 16.Slayden, R. A., and C. E. Barry III. 2002. The role of KasA and KasB in the biosynthesis of meromycolic acids and isoniazid resistance in Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 82:149-160. [DOI] [PubMed] [Google Scholar]
- 17.Slayden, R. A., D. L. Knudson, and J. T. Belisle. 2006. Identification of cell cycle regulators in Mycobacterium tuberculosis by inhibition of septum formation and global transcriptional analysis. Microbiology 152:1789-1797. [DOI] [PubMed] [Google Scholar]
- 18.Slayden, R. A., R. E. Lee, J. W. Armour, A. M. Cooper, I. M. Orme, P. J. Brennan, and G. S. Besra. 1996. Antimycobacterial action of thiolactomycin: an inhibitor of fatty acid and mycolic acid synthesis. Antimicrob. Agents Chemother. 40:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slayden, R. A., R. E. Lee, and C. E. Barry III. 2000. Isoniazid affects multiple components of the type II fatty acid synthase system of Mycobacterium tuberculosis. Mol. Microbiol. 38:514-525. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan, T. J., J. J. Truglio, M. E. Boyne, P. Novichenok, X. Zhang, C. F. Stratton, H.-J. Li, T. Kaur, A. Amin, F. Johnson, R. A. Slayden, C. Kisker, and P. J. Tonge. 2006. High affinity InhA inhibitors with activity against drug-resistant strains of Mycobacterium tuberculosis. ACS Chem. Biol. 1:43-53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.