Abstract
In this study, we systematically investigated the resistance mechanisms to β-lactams, aminoglycosides, and fluoroquinolones of 120 bacteremic strains of Pseudomonas aeruginosa. Pulsed-field gel electrophoresis genotyping showed that 97 of these strains were represented by a single isolate, 10 by 2 and 1 by 3 clonally related isolates, respectively. Seventy-five percent (90 out of 120) of the bacteremic P. aeruginosa strains displayed a significant resistance to one or more of the tested antimicrobials (up to 11 for 1 strain). These strains were found to harbor a great diversity of resistance mechanisms (up to 7 in 1 strain), leading to various levels of drug resistance. Interestingly, 11 and 36% of the isolates appeared to overproduce the MexAB-OprM and MexXY-OprM efflux systems, respectively. Altogether, our results show that P. aeruginosa may accumulate intrinsic (overproduction of cephalosporinase AmpC, increased drug efflux, fluoroquinolone target mutations, and deficient production of porin OprD) and exogenous (production of secondary β-lactamases and aminoglycoside-modifying enzymes) resistance mechanisms without losing its ability to generate severe bloodstream infections. Consequently, clinicians should be aware that multidrug-resistant P. aeruginosa may remain fully pathogenic.
Pseudomonas aeruginosa is a well-known nosocomial pathogen responsible for a wide range of mild to severe infections. Naturally resistant to many antimicrobial agents used in the hospital, this bacterium has the distinctive capacity via multiple mechanisms to become resistant to virtually all the antibiotics available commercially (12, 37). For example, significant resistance to β-lactams may arise from stable up-regulation of the intrinsic cephalosporinase AmpC, acquisition of transferable β-lactamases, increased drug efflux (MexAB-OprM, MexCD-OprJ, and MexXY-OprM systems), or outer membrane impermeability (alteration of porin OprD) (21, 22). Resistance to aminoglycosides may involve aminoglycoside- or 16S RNA-modifying enzymes, as well as the MexXY-OprM efflux pump (26). Similarly, fluoroquinolone resistance may be due to mutations in target genes (gyrA, gyrB, parC, and parE) or to drug efflux systems (MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM) (19, 27).
Despite an abundant literature, little is known about the prevalence of these different mechanisms among clinical strains of P. aeruginosa, how they contribute to drug resistance levels (see reference 7 for a recent review), and whether they influence bacterial virulence. For instance, recent data on in vitro mutants have suggested that the virulence of P. aeruginosa might be reduced when the Mex efflux systems are overexpressed (11, 13, 20, 29, 32).
In this study, we demonstrate that P. aeruginosa may accumulate numerous mechanisms of resistance to β-lactams, aminoglycosides, and fluoroquinolones while involved in bloodstream infections.
MATERIALS AND METHODS
Bacterial strains.
One hundred twenty strains of P. aeruginosa corresponding to the initial isolates of distinct bacteremic episodes were prospectively collected between 1999 and 2004 in six French teaching hospitals that are members of the GESPA group (Groupe d'Etude des Septicémies à Pseudomonas aeruginosa). The genotypic relatedness of these isolates was investigated by pulsed-field gel electrophoresis (CHEF-DR III; Bio-Rad, Ivry sur Seine, France) of DraI-macrorestricted genomic DNA as described previously (31). According to consensual guidelines (33), two isolates were considered clonally related when their pulsed-field gel electrophoresis profiles exhibited less than three bands of difference. The well-characterized mutants PT629 and MutGR-1 from wild-type reference strain PAO1 were used as positive controls in reverse transcription-PCR (RT-PCR) experiments for the identification of gain-of-efflux mutants overexpressing pumps MexAB-OprM and MexXY-OprM, respectively (23).
Drug susceptibility testing.
The MICs of selected antibiotics were determined with the conventional macrodilution technique in Mueller-Hinton agar with calibrated concentrations of divalent cations (MHA; BBL, Cockeysville, MD) (2). Strains were classified as “susceptible,” “intermediate,” or “resistant” according to the CLSI breakpoints (25).
Analysis of β-lactam resistance mechanisms.
The β-lactamase content of the selected strains was first analyzed by isoelectrofocusing (pI) (14) and then confirmed by gene sequencing with consensus primers targeting the tem, shv, and oxa genes (1, 4, 30). In addition, the activities of intrinsic β-lactamase AmpC were quantified spectrophotometrically with the chromogenic substrate nitrocefin (14). Resistant isolates exhibiting basal (uninduced) AmpC activities at least fourfold higher than that of wild-type strain PAO1 (20 nmol/min/mg) were considered to be cephalosporinase-derepressed mutants (21). As expected, the susceptibility of all these mutants to β-lactams (except carbapenems) was partially or completely restored in the presence of the AmpC inhibitor cloxacillin at 1,000 μg/ml (data not shown). The production of the porin OprD was assessed for deficiency by RT-PCR targeting the oprD gene (9) in those isolates with reduced susceptibility to imipenem (MIC, ≥8 μg/ml; n = 21).
Analysis of aminoglycoside resistance mechanisms.
Genes encoding aminoglycoside-modifying enzymes were identified by PCR. Specific sequences of sense and antisense primers were chosen within the nucleotide sequences of the following widely distributed genes in clinical P. aeruginosa: aac(6′)-Ib (5′-TTGCAATGCTGAATGGAGAG-3′ and 5′-CGTTTGGATCTTGGTGACCT-3′), ant(2″)-Ia (5′-GAGCGAAATCTGCCGCTCTGG-3′ and 5′-CTGTTACAACGGACTGGCCGC-3′), and aac(3)-Ia (5′-ACCTACTCCCAACATCAGCC-3′ and 5′-ATATAGATCTCACTACGCGG-3′) (35). Strains showing a moderate resistance to all the aminoglycosides tested, including the enzyme-recalcitrant compounds apramycin and fortimicin, were considered to be harboring at least one nonenzymatic resistance mechanism to these antibiotics (i.e., not involving the production of aminoglycoside-modifying enzymes).
Characterization of gain-of-efflux mutants.
The constitutive up-regulation of the efflux pumps MexAB-OprM and MexXY-OprM was assessed in the 120 clinical strains by (i) semiquantitative estimation of proteins MexB, OprM, and MexY on membrane extracts by Western blotting and (ii) RT-PCR determination of the transcriptional levels of genes mexB and mexY, as reported previously (9, 15, 17). MexXY-OprM-dependent resistance to aminoglycosides and fluoroquinolones was confirmed by overexpressing the plasmid-borne repressor gene mexZ in 10 randomly selected strains (plasmid pAZ17 [36]) and by measuring the residual resistance levels of these bacteria subsequent to mexXY switch-off (15; data not shown).
QRDR sequencing.
A search for mutations in the quinolone resistance-determining regions (QRDR) of genes gyrA, gyrB, parC, and parE was performed in all strains for which the ciprofloxacin MIC was at least equal to 1 μg/ml, as described previously (14).
RESULTS
Epidemiological data.
One hundred twenty bacteremic isolates of P. aeruginosa corresponding to 108 genotypically different strains were recovered from 119 patients admitted to six French teaching hospitals between 1999 and 2004. One patient experienced two septicemic episodes with the same strain at a 43-day interval. Ninety-seven of the collected strains were represented by a single isolate, 10 strains by 2 isolates, and 1 strain by 3 clonally related isolates, respectively. No hospital-to-hospital spread of strains was noted during the survey. Concordant with other data in the literature (8), the 30-day mortality rate after the initial bacteremic episode (arbitrarily defined as the date of the first positive blood culture) was quite high (32%) in this series, with 39% of these deaths directly attributable to bacteremia. The infections were associated with severe underlying conditions, such as leukemias (21%), solid tumors (20%), concurrent infectious diseases (12%), or chronic lung diseases (10%).
Antibiotic resistance.
The rates of susceptibility of the bacteremic isolates to 11 antipseudomonal antibiotics used in French hospitals are shown in Table 1. Piperacillin-tazobactam, ceftazidime, and amikacin were the most-frequently active agents (≥90% susceptible strains). According to the CLSI breakpoints, only 37% (45 of 120) of the isolates were susceptible to all the products tested, with many isolates (63%) exhibiting intermediate or high resistance to at least one antibiotic (up to 9 for three strains) (Fig. 1). From a microbiological viewpoint, as many as 75% (n = 90) of the bacteremic P. aeruginosa isolates displayed a significant resistance (fourfold or greater increase in the MIC of a given antibiotic compared with that for wild-type reference strain PAO1) to one or more of the tested antimicrobials (up to 11 for 1 strain) (Fig. 1). To get an insight into the resistance mechanisms prevailing in these strains, we therefore analyzed several determinants known to increase the MICs of β-lactams, aminoglycosides, and/or fluoroquinolones at least fourfold.
TABLE 1.
Susceptibility rates of the 120 bacteremic isolates of P. aeruginosa
| Antibiotic | MIC range (μg/ml) | Susceptibility rate (%)a
|
MIC50 (μg/ml) | MIC90 (μg/ml) | ||
|---|---|---|---|---|---|---|
| S | I | R | ||||
| Piperacillin-tazobactamb | <0.5-256 | 95.0 | 5.0 | 4 | 64 | |
| Ceftazidime | 0.5-64 | 90.0 | 6.7 | 3.3 | 2 | 8 |
| Imipenem | 0.5-64 | 82.5 | 6.7 | 10.8 | 2 | 16 |
| Cefepime | 1-64 | 81.7 | 15.0 | 3.3 | 4 | 16 |
| Aztreonam | <0.5-64 | 75.8 | 17.5 | 6.7 | 4 | 16 |
| Ticarcillin | 4->256 | 77.5 | 11.7 | 10.8 | 32 | 256 |
| Amikacin | ≤0.5-128 | 93.3 | 5.0 | 1.7 | 4 | 16 |
| Tobramycin | 0.12->64 | 77.5 | 0.8 | 21.7 | 1 | >64 |
| Netilmicin | 0.25->128 | 56.7 | 28.3 | 15.0 | 8 | 32 |
| Gentamicin | ≤0.25-128 | 53.3 | 21.7 | 25.0 | 4 | 128 |
| Ciprofloxacin | 0.06-128 | 69.2 | 2.5 | 28.3 | 0.25 | 32 |
As defined by the CLSI breakpoints. S, susceptible; I, intermediate; R, resistant.
No intermediate category has been defined for this antibiotic by the CLSI.
FIG. 1.
Multidrug resistance in the bacteremic P. aeruginosa isolates. White bars represent the percentage of isolates exhibiting intermediate susceptibility or resistance (CLSI breakpoints) to a given number of antibiotics. Black bars refer to a microbiological definition of resistance (MIC, at least fourfold that of the reference strain PAO1).
Resistance to β-lactams.
Analysis of the 81 isolates showing an increased resistance to β-lactams (as microbiologically defined above) revealed a complex situation involving the overexpression of active efflux systems (MexAB-OprM and/or MexXY-OprM), stable up-regulation of the intrinsic cephalosporinase AmpC, the production of various secondary β-lactamases (PSE-1, TEM-2, or OXA-2), and/or defective production of the carbapenem-specific porin OprD (Table 2). No determinants of resistance to β-lactams could be identified in 12 bacteria exhibiting low susceptibility to piperacillin-tazobactam (n = 5; MIC, 8 μg/ml versus 2 μg/ml for PAO1) and/or cefepime (n = 8; MIC, 4 μg/ml versus 1 μg/ml for PAO1). The remaining 69 strains were found to individually express one (n = 37), two (n = 22), or three (n = 10) different mechanisms of β-lactam resistance. These mechanisms were more often intrinsic (i.e., resulting from chromosomal mutations; n = 58) than exogenous (i.e., resulting from genetic transfers; n = 11). For instance, up-regulation of the β-lactamase AmpC or efflux pump MexXY-OprM occurred in 23 and 43 isolates, respectively. MexAB-OprM overproducing was less prevalent (n = 13). As reported elsewhere (21), AmpC derepression alone caused a 4- to 16-fold increase in the MICs of most of the β-lactams tested, except imipenem. In comparison, efflux-based resistance was somewhat lower (two- to eightfold MIC increase) and restricted to compounds such as cefepime (for MexXY-OprM-overproducing mutants), ticarcillin, and aztreonam (for MexAB-OprM-overproducing mutants). No carbapenemase or extended-spectrum β-lactamase was detected in this study. An alteration in the production of porin OprD typically led to a mean eightfold increase in resistance to the carbapenem imipenem. In 17 strains, oprD transcripts could not be detected by RT-PCR, while in 4 others, the expression of the gene was reduced (0.39- to 0.55-fold that of PAO1).
TABLE 2.
Mechanisms of resistance to β-lactams detected in the bacteremic P. aeruginosa isolates (n = 69)
| Strain(s) and resistance mechanism(s)a | No. of isolates (no. oprD negative) | Modal MICb (μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|
| FEP | CAZ | ATM | TIC | TZP | IPMc | ||
| Wild-type reference strain PAO1 | 1 | 1 | 4 | 16 | 2 | 1 | |
| Wild-type bacteremic isolates | 39 | 2 | 1 | 4 | 16 | 4 | 1 |
| Resistant bacteremic isolates | |||||||
| XY+ | 19 (4) | 4 | 2 | 4 | 16 | 4 | 8 |
| AmpC+, XY+ | 11 (5) | 16 | 8 | 16 | 128 | 64 | 16 |
| OprD− | 8 (8) | 4 | 2 | 4 | 16 | 4 | 8 |
| AmpC+ | 7 (2) | 8 | 16 | 16 | 64 | 32 | 8-64 |
| PSE-1, XY+ | 5 (1) | 16 | 2 | 8 | >256 | 64 | 64 |
| AmpC+, XY+, PSE-1 | 1 (0) | 16 | 8 | 16 | >256 | 128 | |
| PSE-1 | 2 (0) | 16 | 2-4 | 8-16 | >256 | 128-256 | |
| AmpC+, AB+ | 2 (1) | 8 | 4 | 32 | 128 | 16 | 8 |
| AB+ | 6 (0) | 8 | 4 | 16 | 128 | 16 | |
| AB+, XY+ | 4 (0) | 8 | 2 | 16 | 128 | 8 | |
| AmpC+, XY+, TEM-2 | 1 (0) | 16 | 32 | 16 | >256 | 64 | |
| OXA-2 | 1 (0) | 2 | 2 | 16 | 128 | 8 | |
| AmpC+, AB+, XY+ | 1 (0) | 8 | 16 | 16 | 128 | 64 | |
| XY+, TEM-2 | 1 (0) | 16 | 8 | 8 | >256 | 64 | |
Overproduction of MexXY-OprM (XY+) or MexAB-OprM (AB+) pump; production of PSE-1, TEM-2, or OXA-2 β-lactamase; stable overexpression of cephalosporinase AmpC (AmpC+), and deficiency in OprD (OprD−).
MICs in the intermediate or resistant categories, according to CLSI breakpoints, are in boldface. FEP, cefepime; CAZ, ceftazidime; ATM, aztreonam; TIC, ticarcillin; TZP, tazocillin/piperacillin; IPM, imipenem.
MICs of imipenem are only given for OprD− isolates.
Resistance to aminoglycosides.
Half of the isolates (60 of 120; 50%) displayed a minimum fourfold-increased resistance to one or more aminoglycosides (Table 3). RT-PCR and gene complementation experiments showed that a large proportion of these bacteria (43 of 60; 72%) overproduced the efflux system MexXY-OprM. The production of common transferable modifying enzymes, such as ANT(2″)-I, AAC(6′)-I, and AAC(3′)-I, was detected in 25 strains (42%) that were highly resistant to specific aminoglycosides, among which 19 (76%) overexpressed MexXY-OprM. MexXY-OprM up-regulation increased the MICs of aminoglycosides from two- to fourfold regardless of the simultaneous production of modifying enzymes by the bacteria (Table 3). Concordant with this, the proportion of MexXY-OprM gain-of-efflux mutants was found to increase along with the resistance levels to aminoglycosides (3, 68, 55, and 100% at amikacin MICs of ≤4, 8, 16, and ≥32 μg/ml, respectively). Of note, no mechanism could be identified in 11 isolates showing a modal fourfold-increased resistance to all the aminoglycosides tested, including the enzyme-recalcitrant compounds apramycin and fortimicin (data not shown).
TABLE 3.
Mechanisms of resistance to aminoglycosides detected in the bacteremic P. aeruginosa isolates (n = 60)
| Strain(s) and resistance mechanism(s)a | No. of isolates | Modal MICb (μg/ml)
|
|||
|---|---|---|---|---|---|
| NET | TOB | AMK | GEN | ||
| Wild-type reference strain PAO1 | 4 | 0.5 | 4 | 2 | |
| Wild-type bacteremic isolates | 60 | 4 | 1 | 4 | 4 |
| Resistant bacteremic isolates | |||||
| XY+ | 24 | 16 | 2 | 8 | 8 |
| Nonenzymatic, MexXY-OprM-independent resistance | 11 | 16 | 2 | 8 | 8 |
| XY+, ANT(2″)-I | 10 | 16 | >64 | 8 | 128 |
| XY+, AAC(6′)-I | 5 | >128 | >64 | 32 | 128 |
| ANT(2″)-I | 3 | 8 | 16 | 4 | >128 |
| AAC(6′)-I, ANT(2″)-I | 2 | >128 | 64 | 16 | 128 |
| XY+, AAC(6′)-I, ANT(2″)-I | 2 | >128 | 64 | 16 | 128 |
| XY+, AAC(3)-I | 1 | 16 | 2 | 8 | 128 |
| XY+, AAC(6′)-I, AAC(3)-I | 1 | >128 | >64 | 128 | 128 |
| AAC(6′)-I | 1 | >128 | 32 | 16 | 128 |
Production of aminoglycoside-modifying enzymes ANT(2″)-I, AAC(6′)-I, and AAC(3)-I and overproduction of MexXY-OprM pump (XY+).
MICs in the intermediate or resistant categories according to CLSI breakpoints are in boldface. NET, netilmicin; TOB, tobramicin; AMK, amikacin; GEN, gentamicin.
Resistance to fluoroquinolones.
Not less than 61% (73 of 120) of the bacteremic strains exhibited an increased resistance to ciprofloxacin (MIC, ≥0.25 μg/ml versus 0.06 μg/ml for PAO1). Of the 36 strains displaying a low-level resistance to ciprofloxacin (MIC, 0.25 to 1 μg/ml), 9 (25%) and 13 (36%) overproduced the MexAB-OprM or MexXY-OprM efflux pumps, respectively, with three strains overproducing the two pumps simultaneously. The other 37 isolates were nonsusceptible to ciprofloxacin according to the CLSI breakpoint (MIC, ≥2 μg/ml). The canonical Thr-83→Ile substitution in gyrA was found in 35 of these strains, concomitant with a Ser-80→Leu or Ser-80→Trp substitution in parC for 27 of them. Isolates with a double GyrA/ParC mutation were more resistant (modal ciprofloxacin MIC, 32 μg/ml) than strains displaying a single GyrA mutation (modal MIC, 16 μg/ml). One resistant isolate (MIC, 32 μg/ml) displayed a single mutation in parC associated with MexXY-OprM overproduction, while another (MIC, 8 μg/ml) did not exhibit known resistance mechanisms to fluoroquinolones. No mutation was observed in the QRDRs of gyrB and parE genes. Interestingly, most of these resistant isolates (29 of 37; 78%) turned out to overexpress MexXY-OprM in a proportion that increased with the levels of resistance to ciprofloxacin (60, 70, 94, and 100% of the isolates with MICs of 4 to 8, 16, 32, and 128 μg/ml, respectively). Finally, none of 73 isolates displayed a resistance profile compatible with efflux systems MexCD-OprJ and MexEF-OprN being overproduced (hypersusceptibility to most β-lactams and/or to aminoglycosides) (28). This finding agrees with other data showing the low prevalence of MexCD-OprJ- and MexEF-OprN-overexpressing mutants among French clinical strains of P. aeruginosa (16).
DISCUSSION
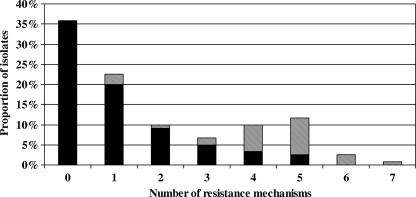
Multidrug resistance in clinical isolates of P. aeruginosa is an increasing threat in hospitals worldwide (22). To our knowledge, an extensive analysis of the mechanisms of resistance to the three major classes of antipseudomonal compounds (β-lactams, aminoglycosides, and fluoroquinolones) in invasive strains of P. aeruginosa has not yet been conducted. This study revealed that a great proportion (75%) of bacteremic strains no longer exhibited a wild-type profile of susceptibility to antimicrobials (MIC, at least fourfold that of the reference strain PAO1). Consistent with this, 64% of the isolates harbored one or several resistance mechanisms (up to six in three strains and seven in one strain) (Fig. 2). The observation that these mechanisms are mostly intrinsic reinforces the notion that P. aeruginosa may readily adapt itself to the antibiotic pressure via chromosomal mutations and does not necessarily require the transfer of foreign DNA (22).
FIG. 2.
Accumulation of distinct resistance mechanisms to antibiotics in the 120 bacteremic P. aeruginosa isolates. The black bars represent the rates of isolates exhibiting only intrinsic resistance mechanism(s) (overexpression of the AmpC β-lactamase, decreased production of porin OprD, up-regulation of efflux systems MexAB-OprM and/or MexXY-OprM, nonenzymatic resistance to aminoglycosides independent of MexXY-OprM, and mutation[s] in the QRDRs). The hatched bars represent the rates of isolates showing both intrinsic and exogenous (production of transferable β-lactamases and aminoglycoside-modification enzymes) mechanisms of resistance.
Such an accumulation of independent mechanisms allows individual strains to become resistant to a large range of antibiotics (complementary effects of the mechanisms) and to survive the most-potent drugs (cooperative effects). For example, the simultaneous overexpression of the cephalosporinase AmpC and efflux system MexXY-OprM in the bacteremic isolates (9%) resulted in resistance to many β-lactams, including the stable third-generation cephalosporin β-lactamase cefepime, as well as to fluoroquinolones and aminoglycosides. Up-regulation of MexXY-OprM reinforced the resistance to aminoglycosides conferred by modifying enzymes (Table 3) and the resistance to fluoroquinolones due to mutations in the QRDRs. As previously noted, synergistic interactions between efflux pumps and β-lactamases were less evident (24).
Interestingly, 15 of the 45 isolates susceptible to all the tested antibiotics (as defined by the CLSI breakpoints) demonstrated low levels of resistance to at least one antibiotic. Ten out of these 15 exhibited a low level of resistance to ciprofloxacin (MIC, 0.25 to 0.5 μg/ml). According to pharmacokinetic/pharmacodynamic studies, a moderate resistance to fluoroquinolones is associated with a dramatic decrease in the target attainment rates that predict therapeutic outcome (10, 18). In some strains, the mechanisms responsible for the low-level resistance to fluoroquinolones (n = 14) and to aminoglycosides (n = 11) could not be characterized. Alteration of the lipopolysaccharide, production of Qnr proteins, and/or decreased uptake of aminoglycosides in these bacteria are currently being investigated (26, 34). Whether these low-level-resistance mechanisms may favor the emergence of secondary mutants with stronger resistance to fluoroquinolones or aminoglycosides needs to be confirmed (3).
Recently, cellular and animal models of infection have suggested that, when overexpressed, Mex pumps might negatively impact the virulence in P. aeruginosa (11, 13, 20, 29, 32). In apparent contrast with these studies, our data show that MexAB-OprM or MexXY-OprM are frequently overproduced in bacteremic strains (11% and 36% of the isolates, respectively). A trivial explanation could be that, in the clinical setting, gain-of-efflux mutants may recover their fitness or virulence by compensatory mutations. Supporting this notion, it has been demonstrated that, while the acquisition of resistance determinants in pathogenic bacteria usually leads to a decreased virulence (5), secondary mutations may easily restore the initial bacterial fitness (5, 6). Of interest, the crude and attributable mortalities in this series were not correlated with the number and nature of the resistance mechanisms accumulated by the bacteremic isolates (data not shown).
In conclusion, this study demonstrates that P. aeruginosa strains with multiple drug resistance mechanisms remain invasive and can cause severe infections.
Acknowledgments
Financial support for the work was provided by GlaxoSmithKline.
We are grateful to Christiane Bailly, Sandra Rochey, Adeline Carmille, and Barbara Dehecq for their excellent technical assistance. We also thank Naomasa Gotoh for providing the MexB antiserum.
Footnotes
Published ahead of print on 6 August 2007.
REFERENCES
- 1.Babini, G. S., and D. M. Livermore. 2000. Are SHV β-lactamases universal in Klebsiella pneumoniae? Antimicrob. Agents Chemother. 44:2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balows, A., W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy. 1991. Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, DC.
- 3.Baquero, F. 2001. Low-level antibacterial resistance: a gateway to clinical resistance. Drug Resist. Updates 4:93-105. [DOI] [PubMed] [Google Scholar]
- 4.Bert, F., C. Branger, and N. Lambert-Zechovsky. 2002. Identification of PSE and OXA β-lactamase genes in Pseudomonas aeruginosa using PCR-restriction fragment length polymorphism. J. Antimicrob. Chemother. 50:11-18. [DOI] [PubMed] [Google Scholar]
- 5.Björkman, J., D. Hughes, and D. I. Andersson. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Björkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479-1482. [DOI] [PubMed] [Google Scholar]
- 7.Bonomo, R. A., and D. Szabo. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43:S49-S56. [DOI] [PubMed] [Google Scholar]
- 8.Diekema, D. J., S. E. Beekmann, K. C. Chapin, K. A. Morel, E. Munson, and G. V. Doern. 2003. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J. Clin. Microbiol. 41:3655-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumas, J.-L., C. Delden, K. Perron, and T. Köhler. 2006. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol. Lett. 254:217-225. [DOI] [PubMed] [Google Scholar]
- 10.Dupont, P., D. Hocquet, K. Jeannot, P. Chavanet, and P. Plésiat. 2005. Bacteriostatic and bactericidal activities of eight fluoroquinolones against MexAB-OprM-overproducing clinical strains of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 55:518-522. [DOI] [PubMed] [Google Scholar]
- 11.Evans, K., L. Passador, R. Srikumar, E. Tsang, J. Nezezon, and K. Poole. 1998. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 180:5443-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falagas, M. E., and S. K. Kasiakou. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40:1333-1341. [DOI] [PubMed] [Google Scholar]
- 13.Hirakata, Y., R. Srikumar, K. Poole, N. Gotoh, T. Suematsu, S. Kohno, S. Kamihira, R. E. Hancock, and D. P. Speert. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 196:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hocquet, D., X. Bertrand, T. Köhler, D. Talon, and P. Plésiat. 2003. Genetic and phenotypic variations of a resistant Pseudomonas aeruginosa epidemic clone. Antimicrob. Agents Chemother. 47:1887-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hocquet, D., P. Nordmann, F. El Garch, L. Cabanne, and P. Plésiat. 2006. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hocquet, D., M. Roussel-Delvallez, J. D. Cavallo, and P. Plésiat. 2007. MexAB-OprM- and MexXY-overproducing mutants are very prevalent among clinical strains of Pseudomonas aeruginosa with reduced susceptibility to ticarcillin. Antimicrob. Agents Chemother. 51:1582-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hocquet, D., C. Vogne, F. El Garch, A. Vejux, N. Gotoh, A. Lee, O. Lomovskaya, and P. Plésiat. 2003. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47:1371-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jumbe, N., A. Louie, R. Leary, W. Liu, M. R. Deziel, V. H. Tam, R. Bachhawat, C. Freeman, J. B. Kahn, K. Bush, M. Dudley, M. H. Miller, and G. L. Drusano. 2003. Application of a mathematical model to prevent in vivo amplification of antibiotic-resistant bacterial populations during therapy. J. Clin. Investig. 112:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Köhler, T., and J. C. Pechère. 1998. Bacterial resistance to quinolones: mechanisms and clinical implications, p. 117-142. In V. T. Andriole (ed.), The quinolones, 2nd ed. Academic Press, San Diego, CA.
- 20.Linares, J. F., J. A. Lopez, E. Camafeita, J. P. Albar, F. Rojo, and J. L. Martinez. 2005. Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187:1384-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 23.Llanes, C., D. Hocquet, C. Vogne, D. Bénali-Baitich, C. Neuwirth, and P. Plésiat. 2004. Clinical strains of Pseudomonas aeruginosa overproducing simultaneously MexAB-OprM and MexXY efflux pumps. Antimicrob. Agents Chemother. 48:1797-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakae, T., A. Nakajima, T. Ono, K. Saito, and H. Yoneyama. 1999. Resistance to β-lactam antibiotics in Pseudomonas aeruginosa due to interplay between the MexAB-OprM efflux pump and β-lactamase. Antimicrob. Agents Chemother. 43:1301-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 2003. Method for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 26.Poole, K. 2005. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 44:2233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poole, K. 2002. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa, p. 201-231. In I. T. Paulsen and K. Lewis (ed.), Microbial multidrug efflux. Horizon Scientific Press, Wymondham, United Kingdom.
- 29.Sanchez, P., J. F. Linares, B. Ruiz-Diez, E. Campanario, A. Navas, F. Baquero, and J. L. Martinez. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 50:657-664. [DOI] [PubMed] [Google Scholar]
- 30.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 75:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talon, D., V. Cailleaux, M. Thouverez, and Y. Michel-Briand. 1996. Discriminatory power and usefulness of pulsed-field gel electrophoresis in epidemiological studies of Pseudomonas aeruginosa. J. Hosp. Infect. 32:135-145. [DOI] [PubMed] [Google Scholar]
- 32.Tan, M.-W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 99:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanhoof, R., H. J. Nyssen, E. Van Bossuyt, and E. Hannecart-Pokorni. 1999. Aminoglycoside resistance in Gram-negative blood isolates from various hospitals in Belgium and the Grand Duchy of Luxembourg. J. Antimicrob. Chemother. 44:483-488. [DOI] [PubMed] [Google Scholar]
- 36.Vogne, C., J. R. Aires, C. Bailly, D. Hocquet, and P. Plésiat. 2004. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 48:1676-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, C. Y., J. S. Jerng, K. Y. Chen, L. N. Lee, C. J. Yu, P. R. Hsueh, and P. C. Yang. 2006. Pandrug-resistant Pseudomonas aeruginosa among hospitalised patients: clinical features, risk-factors and outcomes. Clin. Microbiol. Infect. 12:63-68. [DOI] [PubMed] [Google Scholar]