Abstract
ACC-4, an omega loop mutant (Val211→Gly) of the Hafnia alvei-derived cephalosporinase ACC-1, was encoded by an Escherichia coli plasmid. The genetic environment of blaACC-4 shared similarities with plasmidic regions carrying blaACC-1. Kinetics of β-lactam hydrolysis and levels of resistance to β-lactams showed that ACC-4 was more effective than ACC-1 against expanded-spectrum cephalosporins.
A variety of plasmid-determined β-lactamases belonging to Ambler class C (cephalosporinases) have been identified for members of Enterobacteriaceae, mainly Klebsiella pneumoniae, Escherichia coli, and nontyphoid-causing Salmonella. Based on sequence similarities to species-specific AmpC enzymes, plasmidic cephalosporinases are classified into five evolutionary groups, the most widespread being the group of Citrobacter freundii-originating CMY/LAT variants. The remaining groups include enzymes related to the chromosomal cephalosporinases of Enterobacter cloacae (MIR-1 and ACT-1), Morganella morganii (DHA variants), Hafnia alvei (ACC-1), and Aeromonas spp. (FOX, MOX, and various CMY enzymes) (18).
Plasmid-encoded CMY/LAT cephalosporinases have been repeatedly identified among clinical enterobacteria in Greece (5, 25). A MOX-2-producing K. pneumoniae strain that originated in a Greek hospital has also been reported (21). In a previous screening for acquired bla genes among oxyimino-cephalosporin-resistant E. coli isolates in Athens hospitals, an ACC producer (E. coli EC-3521r) was detected for the first time. E. coli EC-3521r was isolated in 2002 from a urine sample from a patient treated in a general hospital. ACC was encoded by pR3521, a self-transferable plasmid of approximately 80 kb that also mediated production of TEM-1 and a novel carbenicillinase designated SCO-1 (17). It is shown here that the detected ACC-type β-lactamase was a point mutant of ACC-1 with increased activity against expanded-spectrum cephalosporins.
Identification of blaACC-4.
The ACC-encoding plasmid pR3521 was extracted from an E. coli K-12(pR3521) transconjugant clone (17) using a Nucleobond BAC100 kit (Macherey-Nagel, Duren, Germany) and partially digested with Sau3A. Fragments were ligated into the BamHI site of pBCSK(+) (Stratagene, La Jolla, CA), and recombinant plasmids were introduced into E. coli MC4100 (ΔampC) (10) by electroporation. Clones were selected with ampicillin (40 μg/ml) and chloramphenicol (20 μg/ml) and screened for blaACC by PCR using the blaACC-specific primers a-F1 (5′-GACACCGTTGATGACCTGAT-3′) and a-R1 (5′-CACCGAAGCCGTTAGTTGAT-3′) (4). A recombinant plasmid of approximately 5.9 kb (pS18-d) with a blaACC insert was identified. Plasmid pS18-d was purified using a QIAGEN Plasmid Midi kit (QIAGEN, Hilden, Germany), and the blaAAC-containing insert was sequenced on both strands using an ABI 377 sequencer (Applied Biosystems, Foster City, CA). The insert was 2,479 bp long and included a blaACC gene (1,161 bp) that differed from blaACC-1 (1) in a T-to-G transversion (nucleotide [nt] 1,350 in sequence under GenBank accession no. AJ133121), resulting in a Val→Gly substitution at position 211 of ACC-1 (numbering is as in reference 1). This novel ampC variant was designated blaACC-4 (nt 1,995 to 3,155 in the sequence under GenBank accession no. EF504260). The likely secretory signal sequence of the 387-amino-acid-long ACC-4 polypeptide comprised 23 residues. The mature ACC-4 β-lactamase (364 amino acids; molecular weight, 39,673) had an apparent isoelectric point (pI) of 7.8 as determined by analytical isoelectric focusing of cell extracts from E. coli(pS18-d) in ampholine-containing polyacrylamide gels (pH range, 3.5 to 9.5) and using β-lactamases with known pIs as controls (TEM-1, 5.4; TEM-3, 6.3; SHV-3, 7.0; SHV-1, 7.6; SHV-5, 8.2) (data not shown). This pI corresponded to the values reported for ACC-1 and the chromosomal cephalosporinase of H. alvei (1, 7).
Genetic environment of blaACC-4.
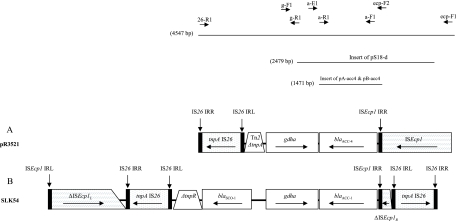
The regions flanking blaACC-4 were determined by sequencing of the entire insert of pS18-d, as well as PCR mapping of pR3521 with primers used for the characterization of ACC-1-encoding plasmids (4) and sequencing of the respective amplicons (Fig. 1). An intact glutamate dehydrogenase gene (GDHA gene) of 915 bp was located downstream of blaACC-4 in the opposite orientation. The remaining left-hand sequence comprised a 175-bp segment homologous to an internal part of tnpA from Tn2 (GenBank accession no. AY123253) and an IS26 element. At an 87-bp distance upstream of blaACC-4, a sequence of 1,306 bp corresponding to an ISEcp1 element was identified. A putative promoter for blaACC-4 was located within ISEcp1 as in the ACC-1-encoding plasmids (4). This segment shared extensive homology with the blaACC-1-carrying region of pSLK54, representing a group of similar plasmids spread among K. pneumoniae and Proteus mirabilis in French hospitals (4, 14) (Fig. 1). Additionally, both pR3521 and pSLK54 carried blaSCO-1, a novel carbenicillinase gene initially reported as orf1 (GenBank accession no. AJ870922 and EF104648), as well as blaTEM-1 (14, 17). However, in the blaACC-1-containing sequence from pSLK54, blaSCO-1 was adjacent to the GDHA gene while it was missing from the respective region in pR3521. PCR assays using primers specific for blaSCO-1 combined with primers for blaACC and the GDHA gene were negative, suggesting that the previously characterized blaSCO-1-containing segment (17) was not in the vicinity of blaACC-4 in pR3521. Also, ISEcp1 of pR3521, unlike that of pSLK54, was not interrupted by IS26 (Fig. 1). It is probable that pR3521 and the group of pSLK54-like plasmids were derived from a common ACC-encoding replicon that diverged through insertion sequence-mediated rearrangements. Association of blaSCO-1 with IS26 in pR3521 as described previously (17) is consistent with this hypothesis.
FIG. 1.
Structures of a blaACC-4-containing segment of plasmid pR3521 (A) and the respective region of the ACC-1-encoding plasmid pSLK54 (B). Arrows indicate the translational orientation of the genes. A tilted line indicates a truncation of the respective gene end. The insert of pS18-d and the locations of primers used in PCR mapping experiments are shown at the top of panel A. Sizes of the respective nucleotide sequences are also indicated.
Kinetic properties of ACC-4.
E. coli(pS18-d) was used as a source of ACC-4. Mid-log-phase cultures (5 liters of tryptone soy broth supplemented with 100 μg of ampicillin per ml) were harvested by centrifugation, and pellets were resuspended in 50 ml Tris buffer (20 mM; pH 7.5). Bacterial cell suspensions were sonicated, and the cell debris was removed by ultracentrifugation. β-Lactamases were purified by ion exchange chromatography essentially as described for chromosomal AmpC of H. alvei (7). Clarified extracts were loaded onto a Q-Sepharose column, and the β-lactamase preparations recovered in the flowthrough were dialyzed against 50 mM phosphate buffer (pH 6.8) and then loaded onto an S-Sepharose column. The proteins were eluted with a linear gradient of NaCl (0 to 0.6 M) in 50 mM phosphate buffer (pH 6.8). β-Lactamase-containing fractions were dialyzed against 50 mM phosphate buffer (pH 7.0). The purity of the final preparation was higher than 95% as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Kinetic parameters of penicillin G, piperacillin, cephalothin, cefoxitin, cefotaxime, ceftazidime, and cefepime hydrolysis at 30°C and pH 7.0 were determined by UV spectrophotometry. The respective wavelengths and extinction coefficients were as described previously (19). Results are presented in Table 1. ACC-4 hydrolyzed cephalothin efficiently due to a high kcat value. Hydrolysis of penicillin G and piperacillin was characterized by low kcat and Km values, which is typical for a class C β-lactamase. ACC-4 exhibited unusually high hydrolytic efficiencies for cefotaxime and ceftazidime, which are considered poor substrates for class C β-lactamases. This was apparently due to a significant increase in the kcat value compared to the respective values reported for plasmid-mediated ACC-1 and chromosomal ACC-2 (1, 7). Measurable hydrolysis of cefepime was also noted. Measurable hydrolysis of cefoxitin by ACC-4 was not observed, as has also been reported for ACC-1 and ACC-2 (1, 7).
TABLE 1.
Kinetic parameters of ACC-4 and ACC-1 for various β-lactam substratesa
| Substrate | Value for hydrolysis of substrate by:
|
|||||
|---|---|---|---|---|---|---|
| ACC-4
|
ACC-1
|
|||||
| kcat (s−1) | Km (μM) | kcat/Km (s−1 · μM−1) | Vmaxb | Km (μM) | Vmax/Km | |
| Cephalothin | 96 ± 6 | 30 ± 3.7 | 3.2 | — | — | — |
| Penicillin G | 0.59 ± 0.05 | 3.8 ± 0.4 | 0.16 | — | — | — |
| Piperacillin | 0.13 ± 0.01 | 1.1 ± 0.15 | 0.12 | 0.17 ± 0.05 | 1.4 ± 0.6 | 0.12 |
| Cefoxitin | <0.01 | —c | — | <0.002 | — | — |
| Cefotaxime | 2.7 ± 0.05 | 9.4 ± 1.0 | 0.29 | <0.01 | — | — |
| Ceftazidime | 1.5 ± 0.1 | 15 ± 1.9 | 0.1 | <0.025 | 17 | <0.01 |
| Cefepime | 0.14 ± 0.01 | 73 ± 12 | 0.002 | — | — | — |
Values for ACC-4 are the means of four independent measurements. Values for ACC-1 are from reference 1.
Expressed as nmol of substrate per min per μg of protein.
—, not determined.
Activities of β-lactamase inhibitors against ACC-4 were assessed at 30°C and pH 7.0 by UV spectrophotometry using cephalothin (100 μM) as a reporter substrate. Enzyme-inhibitor mixtures were preincubated for 5 min before the addition of cephalothin. Results were expressed as 50% inhibitory concentrations (IC50s). Clavulanic acid, sulbactam, and tazobactam exhibited weak inhibitory activities against ACC-4 (IC50s were >500, 250, and 171 μM, respectively). The most potent inhibitors, in descending order, were cloxacillin (IC50, 0.026 μM), aztreonam (IC50, 0.032 μM), and Ro 48-1220 (IC50, 10.3 μM). The inhibitory profile of ACC-4 was characteristic of a class C β-lactamase.
Resistance conferred by ACC-4.
A DNA fragment of 1,471 bp comprising the blaACC-4 gene and its promoter sequence (nt 1,952 to 3,422 in sequence under GenBank accession no. EF504260) was produced by PCR using pS18-d as a template and the primers ecp-F2 (5′-GTTGCTCTGTGGATAACTTG-3′) and a-E1 (5′-ACTCAACATATCGCCTCTCC-3′) (Fig. 1). The fragment was directly ligated into the polycloning site of a Topo TA vector (Invitrogen, Carlsbad, CA). The resulting plasmid was utilized to construct blaACC-1 by site-directed mutagenesis using a mutagenesis kit (Stratagene) and the mutagenic primers MT-1 (5′-AGCCAGTGCACGTGAATATGGAGAT-3′) and MT-2 (5′-ATCTCCATATTCACGTGCACTGGCT-3′) (boldface and underlining indicates nucleotides different from those in the original sequence). For an isogenic comparison of ACC-4 and ACC-1, the similar blaACC-containing segments were cloned into the EcoRI sites of the plasmid vectors pACYC184 (low copy number) and pBCSK(+) (high copy number). Identity and orientation of the inserts were confirmed by sequencing. Recombinant plasmids were introduced into E. coli MC4100, and the MICs of β-lactams were determined by using an agar dilution technique. The pBCSK-derived plasmids, pB-acc4 and pB-acc1, mediated higher resistance levels to most β-lactams tested than the respective pACYC184 recombinants (pA-acc4 and pA-acc1), likely reflecting differences in the blaACC copy number, as also indicated by a six- to sevenfold increase in specific activity against cephalothin. Differences in the β-lactam resistance levels between ACC-4- and ACC-1-producing strains were in line with the hydrolysis data (Table 2). MICs of cefuroxime, cefotaxime, ceftazidime, and aztreonam for the ACC-4 producers were consistently higher than those observed for the respective ACC-1-producing E. coli strains. ACC enzymes conferred similar levels of resistance to the remaining β-lactams, except piperacillin and piperacillin-tazobactan, which were slightly more effective against ACC-4 producers. MICs of cefoxitin, cefepime, and imipenem were low, which is typical for E. coli strains expressing an ACC-type cephalosporinase (1, 7, 14), and were not affected by the moderately increased production of the enzymes mediated by the pBCSK-derived recombinant plasmids (Table 2).
TABLE 2.
β-Lactam susceptibilities of E. coli strains producing ACC-4 and ACC-1 under isogenic conditions
| β-Lactam | MIC (μg/ml) of drug for E. coli MC4100 clones harboring plasmidb:
|
|||||
|---|---|---|---|---|---|---|
| pA-acc4 | pA-acc1 | pACYC184 | pB-acc4 | pB-acc1 | pBCSK | |
| Amoxicillin | >512 | >512 | 4 | >512 | >512 | 4 |
| Piperacillin | 32 | 64 | 0.5 | 128 | 256 | 0.5 |
| Piperacillin + TZa | 16 | 32 | 0.5 | 32 | 64 | 0.5 |
| Cephalothin | >512 | >512 | 2 | >512 | >512 | 2 |
| Cefaclor | >512 | >512 | 2 | >512 | >512 | 2 |
| Cefoxitin | 4 | 4 | 2 | 4 | 4 | 2 |
| Cefotetan | 1 | 1 | 0.5 | 1 | 1 | 0.5 |
| Cefuroxime | >512 | 128 | 1 | >512 | 128 | 1 |
| Ceftazidime | 256 | 32 | 0.25 | 512 | 128 | 0.25 |
| Cefotaxime | 32 | 8 | ≤0.06 | 64 | 16 | ≤0.06 |
| Ceftriaxone | 32 | 16 | 0.12 | 128 | 32 | 0.12 |
| Aztreonam | 2 | 0.5 | ≤0.06 | 2 | 1 | ≤0.06 |
| Cefepime | 0.12 | 0.12 | ≤0.06 | 0.25 | 0.12 | ≤0.06 |
| Imipenem | 0.12 | 0.12 | 0.12 | 0.12 | 0.25 | 0.12 |
TZ, tazobactam at a fixed concentration of 4 μg/ml.
Specific activities (units of activity against cephalothin [1 U corresponded to hydrolysis of 1 μmol of substrate/min/mg of protein]) were as follows: for pA-acc4, 6.3; for pA-acc1, 9.7; for pB-acc4, 43.1; for pB-acc1, 58.8; for pACYC184 and pBCSK, not determined.
Conclusions.
An ACC-1-producing K. pneumoniae strain was first isolated in Germany in 1997 (1). Subsequently blaACC-1-positive enterobacteria were detected in Tunisia, France, Spain, and The Netherlands (2, 6, 8, 11, 13, 14, 16, 18, 22). Isolation of an ACC-producing strain in Greece, as well, indicates an ongoing spread of blaACC mostly in the Mediterranean region due at least in part to an epidemic plasmid.
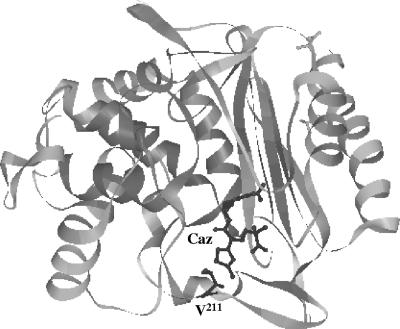
β-Lactam hydrolysis and susceptibility data indicated significant differences between ACC-4 and ACC-1. Val211 is common among intrinsic and acquired AmpCs from members of Enterobacteriaceae, with the exception of Providencia stuartii AmpC, which nevertheless also contains an amino acid with an aliphatic side chain (Leu211) and two FOX variants, FOX-4 and FOX-7, containing Ala211 (GenBank accession no. Y17315, AJ277535, and AJ703796, respectively). Val211 is also encountered in the majority of the AmpC enzymes from Pseudomonas, Aeromonas, and Acinetobacter, while Gly211 has not been observed before. Val211 in ACC-1 is most probably located in the Ω loop, as indicated by a Clustal W alignment (not shown) as well as the extensive similarities to other enterobacterial AmpCs with known structures (e.g., 40% identity and 64% similarity with AmpC from E. coli). Mutational alterations, including substitutions and insertions in this domain in natural and in vitro-selected enterobacterial AmpC variants, facilitate hydrolysis of expanded-spectrum cephalosporins, mainly by increasing the kcat values, as also observed for ACC-4 (9, 12, 15, 23, 24, 26). Such functional changes have been associated with a wider active site and/or more flexibility of the Ω loop (3, 9, 26). Notably, the low hydrolytic efficiencies of AmpCs against expanded-spectrum cephalosporins have been partly attributed to a steric clash of the bulky R1 substituent of these substrates with the side chain of Val211 (Fig. 2). Ω-Loop mutations may alter the position of this residue, thus allowing catalytically more-efficient substrate docking (20). The effect of Gly-for-Val211 substitution in ACC as described here is compatible with an important role of Val211 in determining the substrate specificity of AmpC β-lactamases. Additionally, these findings also underscore the role of the Ω loop as a hot spot for extended-spectrum mutations in the H. alvei-originating cephalosporinases.
FIG. 2.
Ribbon diagram of AmpC from E. coli in complex with acylated ceftazidime (Caz) (PDB entry 1IEL) (20), displaying the proximity of the side chain of Val211 to the aminothiazole group of the antibiotic. The image was made using the DeepView/Swiss-Pdb Viewer, version 3.7, available at www.expasy.org/spdbv/.
Nucleotide sequence accession number.
The GenBank accession number of the nucleotide sequence reported in this study is EF504260.
Acknowledgments
The β-lactamase inhibitor Ro 48-1220 was kindly provided by Roche. We thank Dimitra Gianneli for E. coli EC-3521r, Laurent Poirel for E. coli MC4100, and Christos Stergiou for expert advice on protein purification.
Footnotes
Published ahead of print on 30 July 2007.
REFERENCES
- 1.Bauernfeind, A., I. Schneider, R. Jungwirth, H. Sahly, and U. Ullmann. 1999. A novel type of AmpC β-lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob. Agents Chemother. 43:1924-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidet, P., B. Burghoffer, V. Gautier, N. Brahimi, P. Mariani-Kurkdjian, A. El-Ghoneimi, E. Bingen, and G. Arlet. 2005. In vivo transfer of plasmid-encoded ACC-1 AmpC from Klebsiella pneumoniae to Escherichia coli in an infant and selection of impermeability to imipenem in K. pneumoniae. Antimicrob. Agents Chemother. 49:3562-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crichlow, G. V., A. P. Kuzin, M. Nukaga, K. Mayama, T. Sawai, and J. R. Knox. 1999. Structure of the extended-spectrum class C β-lactamase of Enterobacter cloacae GC1, a natural mutant with a tandem tripeptide insertion. Biochemistry 38:10256-10261. [DOI] [PubMed] [Google Scholar]
- 4.Doloy, A., C. Verdet, V. Gautier, D. Decré, E. Ronco, A. Hammami, A. Philippon, and G. Arlet. 2006. Genetic environment of acquired blaACC-1 β-lactamase gene in Enterobacteriaceae isolates. Antimicrob. Agents Chemother. 50:4177-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gazouli, M., L. S. Tzouvelekis, A. C. Vatopoulos, and E. Tzelepi. 1998. Transferable class C β-lactamases in Escherichia coli strains isolated in Greek hospitals and characterization of two enzyme variants (LAT-3 and LAT-4) closely related to Citrobacter freundii AmpC β-lactamase. J. Antimicrob. Chemother. 42:419-425. [DOI] [PubMed] [Google Scholar]
- 6.Girlich, D., A. Karim, C. Spicq, and P. Nordmann. 2000. Plasmid-mediated cephalosporinase ACC-1 in clinical isolates of Proteus mirabilis and Escherichia coli. Eur. J. Clin. Microbiol. Infect. Dis. 19:893-895. [DOI] [PubMed] [Google Scholar]
- 7.Girlich, D., T. Naas, S. Bellais, L. Poirel, A. Karim, and P. Nordmann. 2000. Biochemical-genetic characterization and regulation of expression of an ACC-1-like chromosome-borne cephalosporinase from Hafnia alvei. Antimicrob. Agents Chemother. 44:1470-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasman, H., D. Mevius, K. Veldman, I. Olesen, and F. M. Aarestrup. 2005. Beta-lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56:115-121. [DOI] [PubMed] [Google Scholar]
- 9.Hidri, N., G. Barnaud, D. Decre, C. Cerceau, V. Lalande, J. C. Petit, R. Labia, and G. Arlet. 2005. Resistance to ceftazidime is associated with a S220Y substitution in the omega loop of the AmpC β-lactamase of a Serratia marcescens clinical isolate. J. Antimicrob. Chemother. 55:496-499. [DOI] [PubMed] [Google Scholar]
- 10.Honoré, N., M. H. Nicolas, and S. T. Cole. 1989. Regulation of enterobacterial cephalosporinase production: the role of a membrane-bound sensory transducer. Mol. Microbiol. 3:1121-1130. [DOI] [PubMed] [Google Scholar]
- 11.Makanera, A., G. Arlet, V. Gautier, and M. Manai. 2003. Molecular epidemiology and characterization of plasmid-encoded β-lactamases produced by Tunisian clinical isolates of Salmonella enterica serotype Mbandaka resistant to broad-spectrum cephalosporins. J. Clin. Microbiol. 41:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumura, N., S. Minami, and S. Mitsuhashi. 1998. Sequences of homologous β-lactamases from clinical isolates of Serratia marcescens with different substrate specificities. Antimicrob. Agents Chemother. 42:176-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miró, E., B. Mirelis, F. Navarro, L. Matas, M. Gimenez, and C. Rabaza. 2005. Escherichia coli producing an ACC-1 class C beta-lactamase isolated in Barcelona, Spain. Antimicrob. Agents Chemother. 49:866-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadjar, D., M. Rouveau, C. Verdet, J. L. Donay, J. L. Herrmann, P. H. Lagrange, A. Philippon, and G. Arlet. 2000. Outbreak of Klebsiella pneumoniae producing transferable AmpC-type β-lactamase (ACC-1) originating from Hafnia alvei. FEMS Microbiol. Lett. 187:35-40. [DOI] [PubMed] [Google Scholar]
- 15.Nukaga, M., S. Haruta, K. Tanimoto, K. Kogure, K. Taniguchi, M. Tamaki, and T. Sawai. 1995. Molecular evolution of a class C β-lactamase extending its substrate specificity. J. Biol. Chem. 270:5729-5735. [DOI] [PubMed] [Google Scholar]
- 16.Ohana, S., V. Leflon, E. Ronco, M. Rottman, D. Guillemot, S. Lortat-Jacob, P. Denys, G. Loubert, M. H. Nicolas-Chanoine, J. L. Gaillard, and C. Lawrence. 2005. Spread of a Klebsiella pneumoniae strain producing a plasmid-mediated ACC-1 AmpC β-lactamase in a teaching hospital admitting disabled patients. Antimicrob. Agents Chemother. 49:2095-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papagiannitsis, C. C., A. Loli, L. S. Tzouvelekis, E. Tzelepi, G. Arlet, and V. Miriagou. 2007. SCO-1, a novel plasmid-mediated class A β-lactamase with carbenicillinase characteristics from Escherichia coli. Antimicrob. Agents Chemother. 51:2185-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type beta-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Power, P., M. Galleni, J. A. Ayala, and G. Gutkind. 2006. Biochemical and molecular characterization of three new variants of AmpC β-lactamases from Morganella morganii. Antimicrob. Agents Chemother. 50:962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powers, R. A., E. Caselli, P. J. Focia, F. Prati, and B. K. Shoichet. 2001. Structures of ceftazidime and its transition-state analogue in complex with AmpC β-lactamase: implications for resistance mutations and inhibitor design. Biochemistry 40:9207-9214. [DOI] [PubMed] [Google Scholar]
- 21.Raskine, L., I. Borrel, G. Barnaud, S. Boyer, B. Hanau-Berçot, J. Gravisse, R. Labia, G. Arlet, and M.-J. Sanson-Le-Pors. 2002. Novel plasmid-encoded class C β-lactamase (MOX-2) in Klebsiella pneumoniae from Greece. Antimicrob. Agents Chemother. 46:2262-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhimi-Mahjoubi, F., M. Bernier, G. Arlet, Z. Ben Jemaa, P. Jouve, A. Hammami, and A. Philippon. 2002. Identification of plasmid-encoded cephalosporinase ACC-1 among several enterobacteria (Klebsiella pneumoniae, Proteus mirabilis, Salmonella) issued from a Tunisian hospital (Sfax, 1997-2000). Pathol. Biol. 50:7-11. [DOI] [PubMed] [Google Scholar]
- 23.Tsukamoto, K., R. Kikura, R. Ohno, and T. Sawai. 1990. Substitution of aspartic acid-217 of Citrobacter freundii cephalosporinase and properties of the mutant enzymes. FEBS Lett. 264:211-214. [DOI] [PubMed] [Google Scholar]
- 24.Tsukamoto, K., R. Ohno, M. Nukaga, and T. Sawai. 1992. The effect of amino acid substitution at position 219 of Citrobacter freundii cephalosporinase on extension of its substrate spectrum. Eur. J. Biochem. 207:1123-1127. [DOI] [PubMed] [Google Scholar]
- 25.Tzouvelekis, L. S., E. Tzelepi, and A. F. Mentis. 1994. Nucleotide sequence of a plasmid-mediated cephalosporinase gene (blaLAT-1) found in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 38:2207-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, Z., Y. Yu, J. M. Musser, and T. Palzkill. 2001. Amino acid sequence determinants of extended-spectrum cephalosporin hydrolysis by the class C P99 β-lactamase. J. Biol. Chem. 276:46568-46574. [DOI] [PubMed] [Google Scholar]