Abstract
The colonic microbiota is a major modulator of the mucosal immune system; therefore, its manipulation through supplementation with probiotics may significantly affect the host's immune responses. Since different probiotics seem to exert various effects in vivo, we tested the relevance of the autoaggregation phenotype on the intestinal persistence of lactobacilli and their ability to modulate the host's innate immune responses. After 14 days of diet supplementation, the aggregating strain Lactobacillus crispatus M247 but not aggregation-deficient isogenic mutant MU5 was recovered from the feces and colonic mucosa of mice. This observation was confirmed by strain-specific PCR amplification and by Lactobacillus-specific denaturing gradient gel electrophoresis analysis. Indeed, L. crispatus M247 increased Toll-like receptor 2 (TLR2) mRNA levels, while it reduced TLR4 mRNA and protein levels in the colonic mucosa, whereas MU5 was ineffective. In colonic epithelial cells (CMT-93 cells) L. crispatus M247 but not MU5 induced time-dependent extracellular signal-regulated kinase-1 (ERK1) tyrosine phosphorylation and TLR modulation, which were abolished in the presence of PD98059 (an ERK1 inhibitor). To assess the functional relevance of probiotic-induced TLR modulation, we determined the consequences of L. crispatus preexposure on TLR4 (lipopolysaccharide [LPS]) and TLR2 [Pam3Cys-Ser-(Lys)4] ligand-mediated effects in intestinal epithelial cells. Preexposure to L. crispatus M247 blunted LPS-induced interleukin-6 (IL-6) release and inhibition of CMT-93 migration over a wound edge, whereas it enhanced TLR2-mediated IL-10 up-regulation. In summary, the aggregation phenotype is required for L. crispatus persistence in the colon and for modulation of TLR2/TLR4 expression through an ERK-dependent pathway. We speculate that the aggregation phenotype in L. crispatus M247 is required to temper epithelial cell responsiveness to bacterial endotoxins, which thus affects the evolution of intestinal inflammatory processes.
Humans and animals are born germfree, but soon after birth they are colonized with microorganisms. Within a few days, the mucosae and the skin are the homes to a vast and complex community of microorganisms. Indeed, 400 species are estimated to inhabit the gastrointestinal tract and establish life-long interactions with the host mucosae to influence a variety of activities of paramount relevance, including the function of the mucosal immune system (27). On the other hand, components of the intestinal microbiota possess the potential to damage the mucosa either through toxin release or as a cause of detrimental immune responses. Thus, in a variety of animal models, intestinal inflammation does not occur when mice are raised under germfree conditions unless their bacterial flora is reconstituted (53). In accordance with the complex effects of the colonic flora on the mucosal immune system, changes in the mucosa-associated microbiota have been related to a variety of diseases (53). Indeed, manipulation of the flora colonizing mucosal surfaces by oral supplementation with live bacteria might influence the host's health and has been proposed as a means for the prevention or treatment of a range of diseases, although the mechanism(s) of action of probiotics remains elusive (53). Thus, probiotics can directly suppress the growth of pathogens through the secretion of antimicrobial substances or induce the expression of protective molecules to enhance the mucosal barrier function (56, 59). Furthermore, probiotics modulate the mucosal immune system either directly, affecting immune cell activities, or through the manipulation of the colonic microbiota. However, different probiotics seem to exert various effects on the host, suggesting the existence of distinctive strain properties (20). Despite the growing number of clinical applications, at the moment no phenotypic markers with which probiotic effects can be predicted in vivo are available.
The gut epithelial cells are no longer considered a mechanical barrier to the prevention of microbial invasion, as they directly sense the gut environment and activate a variety of intracellular pathways in response to specific bacterium-derived products (55). A major breakthrough in the understanding of the molecular mechanisms involved in regulating the bacteria-host interaction was the demonstration that immune and nonimmune cells, including intestinal epithelial cells, recognize several microbial products, referred to as “pathogen-associated molecular patterns,” like the lipopolysaccarides (LPSs) of gram-negative bacteria and the peptidoglycan fragments of gram-positive bacteria, through molecules called “pattern recognition receptors” (PRRs) (9). Among the PRRs are the mammalian homologues of Drosophila Toll receptors, referred to a Toll-like receptors (TLRs), which are transmembrane proteins characterized by an extracellular domain able to bind different pathogen-associated molecular patterns (5). Thus, TLR4 is the prototype of the gram-negative bacterial LPS receptor, whereas TLR2 is the main receptor for peptidoglycan fragments and lipoteichoic acid from gram-positive bacteria. Individual TLRs differentially activate distinct signaling events via cofactors and adaptor proteins, leading to the activation and nuclear translocation of transcription factors. These factors modulate the expression of pro- and anti-inflammatory cytokines and chemokines, which regulate the activities of the innate and the adaptive immune responses (7). These events are involved to control host homeostasis, pathogen suppression, and the responses to probiotic ingestion (19, 33, 62).
To exert favorable effects on the host, administered probiotics are supposed to induce intestinal colonization and to manipulate the colonic microbiota (53). However, to draw a comprehensive picture of the colonic and fecal microbiota of humans and animals, traditional culture-based methods are nowadays considered obsolete for the large number of noncultivable microorganisms, whereas several molecular tools allow the identification of strictly anaerobic species, which are usually predominant in the large bowel of mammals (6, 36, 52, 62). Thus, PCR coupled with denaturing gradient gel electrophoresis (DGGE) was recently applied to the study of complex bacterial communities, with a particular focus on the gut microflora (24, 64) and its fluctuations in diseases or following probiotic administration (17, 29, 31).
Since in a previous study we observed that aggregation-deficient Lactobacillus crispatus MU5 was devoid of therapeutic effects in a colitis model, as opposed to wild-type strain M247, which has an aggregation phenotype, we hypothesized that the aggregation phenotype might give the probiotic strains advantages that are relevant to their in vivo effects (13). Thus, in the study described here, we assessed the impact of oral supplementation with two isogenic strains of L. crispatus, spontaneously aggregating strain M247 and aggregation-deficient strain MU5, on the colonic microbiota and the associated effects on the mucosal level of PRRs, with the view that the levels of these receptors contribute to the establishment of the responsiveness of the mucosa-associated immune system to bacterium-derived products and regulate the amplitudes of the inflammatory responses.
MATERIALS AND METHODS
Isolation, characterization, and culture of Lactobacillus crispatus.
Lactobacillus crispatus strain M247 was isolated and characterized as described previously (14). Cells grown in De Man-Rogosa-Sharpe (MRS; Difco) medium appeared to the naked eye as discernible clumps which sediment at the bottom of the tube, leaving the upper part of the medium clear. A spontaneous nonclumping mutant of M247, named MU5, was isolated from the lower aqueous phase during a hydrophobic assessment test based on the water-hexadecane partition assay. Strains were grown in MRS broth or agar at 37°C under microaerophilic conditions (10% CO2 in air [21% O2]).
Preparation of orally administered cultures.
L. crispatus M247 and MU5 were grown in MRS medium at 37°C for 18 h. The cells were harvested by centrifugation at 8,000 rpm for 5 min, washed twice with sterile distilled saline, and finally suspended in GG solution (20% glucose plus 10% glycerol) to obtain a final concentration of 108 CFU per 100 μl of bacterial suspension.
Administration of L. crispatus M247 and MU5 to mice.
BALB/c mice (age, 8 to 10 weeks) purchased from Charles River (Charles River Laboratories, Lecco, Italy) were used in all experiments. The animal studies were approved by the Institutional Animal Care and Use Committee of the University of Padua. The animals, housed in groups of four mice per cage, were randomly divided into three experimental groups of 8 to 12 elements each. The mice received 108 CFU of L. crispatus M247 or MU5, whereas control animals received only GG solution (vehicle). The microorganisms were administered daily intragastrically via a polyethylene cannula in a total volume of 100 μl. During the treatment period the animals had free access to food and water. After 14 days the animals were killed by use of an overdose of anesthesia (xylazine-ketamine; 100 mg/kg of body weight), the abdomen was immediately opened, and the proximal colon was removed. For each mouse a colon sample was frozen in liquid nitrogen for RNA extraction. Full-thickness specimens of the colon were fixed in 4% paraformaldehyde and embedded in paraffin, and 10-μm-thick sections were stained with hematoxylin-eosin for routine histological examination. An adjacent colon segment was placed in a cryoembedding matrix (OCT) and frozen in isopentane at −110°C. In addition, tissue samples were placed in ice-cold RPMI 1640 medium (Gibco, Milan, Italy) and were immediately processed for epithelial cell isolation.
Persistence study.
Fecal samples were collected immediately before the first administration (time zero) and after 7 days and 14 days of M247 and MU5 supplementation. A sample of the colonic mucosa was also collected after 14 days of probiotic supplementation. Fecal pellets and colonic tissue specimens were immediately placed in Amies medium (3) and stored at 4°C until they were processed (within 24 h). Fecal samples were then serially diluted with sterile saline solution, plated onto MRS medium (Difco), and incubated in an anaerobic atmosphere (85% N2, 10% H2, 5% CO2) at 37°C for 48 h. To recover bacteria adherent to the mucosa, colonic tissue samples were washed twice in sterile saline and subjected to hypotonic lysis, and then the debris was collected by centrifugation and seeded onto MRS plates and incubated under anaerobic conditions. White colonies were then replica plated and incubated anaerobically at 37°C for 48 h. The isolated strains were then identified by strain-specific PCR with primers specific for L. crispatus M247 and MU5, as described by Cesena et al. (14).
DNA extraction, PCR amplification, and DGGE analysis.
The DNA was directly extracted from fecal samples by means of a PSPSpin stool kit (Invitrogen, Germany). The eluted DNA was amplified by nested PCR with two different primer pairs. The PCR products obtained by using primers S-D-Bact-0011-a-A-17 and S-G-Lab-0677-a-A-17 (24) were then used as templates in nested PCR with primers S-G-Lab-0159-A-S-20 and S-Univ-0515-a-A-24-GC (31). The cycling conditions were those previously indicated by Heilig et al. (24) and Konstantinov et al. (31). The amplicons were then analyzed by gel electrophoresis and were visualized by ethidium bromide staining.
DGGE analysis was performed with an Ingeny2×2 system (IngenyPhor, Denmark), which was run at 60°C and a voltage of 120 V for 16 h. An 8% polyacrylamide gel with a 30% to 60% urea gradient was loaded with 40 μl of amplified sample and run with 1× TAE (Tris-acetate-EDTA) buffer. The gel was stained with SYBR green dye (Bio-Rad) and was viewed under UV transillumination. The amplified fragments were then excised from the gel, and the DNA was eluted in sterile distilled water after 18 h incubation at 4°C. A small amount of the eluted DNA was submitted to PCR amplification with primers S-G-Lab-0159-A-S-20 and S-Univ-0515-a-A-24-GC, which were described previously (31). The amplicons were purified with a Microclean kit (Microzone, United Kingdom), and the eluted DNA was sequenced with primer S-G-Lab-0159-A-S-20 at CRIBI, University of Padua. The sequences were then analyzed as described by Knarreborg et al. (29).
Isolation of epithelial cells from mouse colon.
Pieces of colon tissue from the control and the L. crispatus-supplemented mice were surgically removed and washed with RPMI 1640 medium to remove mucus and fecal matter and were placed into Hank's balanced salt solution (HBSS; GIBCO BRL) containing Ca2+, Mg2+, and 1 mM dithiothreitol for 20 min at room temperature with occasional agitation. To remove and recover epithelial cells, the specimens of colonic tissues obtained from the three different experimental groups were then cut into small pieces (2 by 2 mm) and placed in HBSS supplemented with 1 mM EDTA but without Ca2+ and Mg2+. The specimens were incubated for 30 min at 37°C with agitation (170 rpm), and then the medium was collected and the epithelial cells were pelleted by centrifugation (2,000 × g for 10 min). Contaminating intraepithelial lymphocytes were removed by centrifugation (2,000 × g for 20 min) through a Percoll gradient (Amersham-Pharmacia, Milan, Italy). The epithelial cells were collected, washed twice in ice-cold phosphate-buffered saline (PBS), and stored at −80°C for subsequent RNA extraction.
Cell culture.
The CMT-93 mouse colonic epithelial cell line was obtained from the European Collection of Cell Cultures (ECACC no. 89111413). The CMT-93 cells were maintained in Dulbecco's modified Eagle medium (GIBCO) supplemented with 10% heat-inactivated fetal calf serum (GIBCO), 100 U/ml penicillin, and 100 μg/ml streptomycin (complete medium). Subconfluent monolayers were trypsinized, resuspended at 106 cells/ml, and seeded in 6- or 12-well tissue culture plates (Costar). At 7 days postconfluence, the medium was removed; and the cells were washed twice in antibiotic-free medium (AFM) and refed fresh AFM alone or AFM containing 108 CFU/ml L. crispatus M247, L. crispatus MU5, or enteropathogenic Escherichia coli ATCC 49106. When it was so indicated, the monolayers were treated with the specific extracellular signal-regulated kinase-1 (ERK1) inhibitor (PD98059; Calbiochem) 30 min before exposure to the bacteria. After 1 h of coincubation with L. crispatus at 37°C, the monolayers were washed three times with AFM and refed with complete medium, and when it was so indicated, the monolayers were treated with TLR2 [Pam3Cys-Ser-(Lys)4 (Pam3CSK4)] or TLR4 (LPS) ligand (10 μg/ml). After an additional 0 to 24 h of incubation at 37°C, the cells were removed with a cell scraper, washed with ice-cold PBS, centrifuged (1,500 × g for 10 min), and used for RNA or protein extraction.
RNA extraction and quantitative real-time RT-PCR analysis.
Samples of the colonic mucosa or purified epithelial cells were placed in 175 μl of SV RNA lysis buffer from the SV total RNA isolation system kit obtained from Promega Corporation (Madison, WI) and homogenized with a Retsch MM300 apparatus (QIAGEN, Milan Italy). The total RNA was then purified according to the manufacturer's protocol, and the contaminating DNA was removed by DNase I digestion. RNA purity was confirmed by assessing the optical density at 260 nm (OD260) and the OD280. Samples (3 μg of total RNA) with OD260/OD280 ratios of between 1.8 and 2 were used to generate randomly primed cDNAs with Moloney murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA).
Real-time quantitative reverse transcription-PCR (RT-PCR) analysis for interleukin-6 (IL-6), IL-1, IL-10, TLR4, and TLR2 mRNAs was performed on an ABI Prism 7700 sequence detector (Applied Biosystems, Milan, Italy). The oligonucleotide primer sequences and PCR conditions used are reported in Table 1. Quantitative RT-PCR analysis was performed with a SYBR green PCR core reagents kit (Applied Biosystems, Milan, Italy), according to the manufacturer's protocol. Standard curves were obtained by amplification of the corresponding cDNAs subcloned into the pGEM-T vector (Promega, Italy). The expression of the target genes was normalized to that of the endogenous control gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
TABLE 1.
PCR primers and conditions used in the study
| Primer sequence
|
Melting temp (°C) | Length (bp) | ||
|---|---|---|---|---|
| Forward | Reverse | |||
| IL-1 | 5′-TCAATGGACAGAATATCAACCAAC-3′ | 5′-CTTTTCCATCTTCTTCTTTGGGAT-3′ | 60 | 200 |
| IL-6 | 5′-CCGGAGAGGAGACTTCACAGA-3′ | 5′-TCCACGATTTCCCAGAGAAC-3′ | 63 | 105 |
| IL-10 | 5′-ATGCAGGACTTTAAGGGTTACTTG-3′ | 5′-ACTGCCTTGCTCTTATTTTCACAG-3′ | 62 | 206 |
| TLR4 | 5′-TGAGATTGCTCAAACATGGC-3′ | 5′-CGAGGCTTTTCCATCCAATA-3′ | 55 | 213 |
| TLR2 | 5′-CAGCTGGAGAACTCTGACCC-3′ | 5′-CAAAGAGCCTGAATGGGGAG-3′ | 61 | 205 |
| GAPDH | 5′ACTCCACTCACGGCAAATTC-3′ | 5′-TCTCCATGGTGGTGAAGACA-3′ | 47 | 177 |
Immunoprecipitation and Western blotting.
To extract total proteins from CMT-93 cells following the treatments, the cells were washed twice with ice-cold PBS and then lysed (45 min on ice) with nondenaturing RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, 0.25% sodium deoxycholate, 0.1% Nonidet P-40, 100 μM NaVO4, 1 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). Particulate material was removed by centrifugation (15,000 × g for 5 min at 4°C), the supernatants were collected, and the protein concentrations were determined by the Bradford method (Pierce, Cramblington, United Kingdom). To assess ERK1 phosphorylation following L. crispatus exposure, cell lysates (2 mg/ml) were incubated with a rabbit anti-ERK1 polyclonal antibody (10 μg/mg cell lysate; Santa Cruz Biotechnology, Santa Cruz, CA) for 2 h at 4°C. Then, protein A-agarose (Santa Cruz Biotechnology) was added and the mixture was incubated for 1 h at 4°C. Beads were washed twice by centrifugation (20 s, 12,000 × g) with ice-cold RIPA buffer, followed by one wash with ice-cold PBS, and were then boiled in 25 μl of sample loading buffer (62.5 mM Tris, pH 6.8, 10% glycerol, 2% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol, and 0.1% bromophenol blue). The immunoprecipitated proteins were then fractionated on an SDS-polyacrylamide gel and transferred to and immobilized on a nitrocellulose membrane. To determine the TLR2 expression level, 40 μg of total proteins was boiled in loading buffer and then fractionated on an SDS-polyacrylamide gel and transferred to and immobilized on a nitrocellulose membrane. The membranes were blocked overnight at 4°C in 5% skim milk in PBS containing 0.05% Tween 20 and were then incubated for 2 h with the proper antibody (anti-TLR2, anti-phosphotyrosine pY99; Santa Cruz Biotechnology). Bound antibody was then detected by incubating the nitrocellulose membrane with horseradish peroxidase-conjugated donkey anti-goat (for anti-TLR2) immunoglobulin G (IgG) antibody (Santa Cruz Biotechnology), and the immunocomplexes were visualized by using enhanced chemiluminescence Western blot analysis detection reagents (Santa Cruz Biotechnology). The membranes were photographed with a VersaDoc imaging system (Bio-Rad), and the images were digitally stored with Quantity One (Bio-Rad) software.
Immunohistochemistry.
Immunohistochemistry was performed with colonic tissue sections (10 μm thick) obtained from control and L. crispatus-supplemented mice. The frozen sections were fixed in methanol (5 min at −20°C) and then washed twice (5 min each) in Tris-buffered saline (TBS), and nonspecific binding was blocked by incubation with 2% donkey serum in TBS for 20 min. The sections were then incubated with properly diluted primary antibody (rabbit polyclonal anti-TLR4 or goat anti-TLR2 antibody; Santa Cruz Biotechnology) for 2 h at 22°C. Nonbound antibody was removed by extensive washes with TBS. Immunocomplexes were detected with goat Alexa Fluo 488-labeled anti-rabbit IgG (Invitrogen Corporation, Italy) or a rabbit Alexa Fluo 488-labeled anti-goat IgG (Invitrogen Corporation, Italy). The sections were then washed, mounted, analyzed, and photographed with a Leica TCSNT/SP2 confocal microscope (×63 objective). The images were digitally stored by using Leica software and were then elaborated by using a graphics program (Adobe).
IL-6 release.
Following 1 h incubation with AFM alone or AFM supplemented with 108 CFU/ml L. crispatus M247 or MU5, the CMT-93 cell monolayers were exposed to LPS (10 μg/ml) for 6 h. Then, the culture medium was collected, centrifuged (2,000 rpm for 10 min) to remove the deattached cells, and stored at −20°C. IL-6 release was measured by a commercially available enzyme-linked immunosorbent assay (Biosource, Camarillo, CA), following the manufacturer's protocol. The results were expressed as ng/ml.
Migration (restitution) assays.
Assays for wound healing were carried out as described previously (34). The CMT-93 cells dissociated with trypsin-EDTA were seeded onto microscope coverslips and grown in complete medium. At 7 days postconfluence, the monolayers were washed and then incubated with AFM alone or AFM supplemented with 108 CFU/ml of L. crispatus M247 or MU5. One hour later, the medium was removed and the cells were washed with sterile PBS and incubated with complete medium. The monolayers were incubated for an additional 2 h at 37°C in 5% CO2 before linear wounds were made with a sterile razor blade. Then, the cells were incubated in AFM alone or AFM supplemented with LPS (10 μg/ml) for an additional 24 h. Finally, the monolayers were washed in cold PBS, fixed in buffered 2% paraformaldehyde for 5 min, and then mounted on microscope slides. Migration was assessed in a blinded fashion by determining the number of CMT-93 cells across the wound border in a defined wound area by taking photomicrographs at a fivefold magnification with a Leica DM-LB inverted microscope connected to a digital Leica DC-100 camera. The experiments were performed in triplicate, and at least 10 wound areas were used to quantify the migration.
Statistical analysis.
The data are expressed as means ± standard errors (SEs). Statistical analysis was performed by using a t test for unpaired samples. Statistical significance was considered for a P value of <0.05.
RESULTS
Persistence of L. crispatus M247 in feces and adherence to colonic mucosa of mice.
Fecal samples from each animal were collected before the treatment with L. crispatus was started and after 7 and 14 days of daily supplementation with 108 CFU of either L. crispatus M247 or MU5. L. crispatus M247 or MU5 was not identified in the feces of any animal at the beginning of the experiment. Viable M247 cells were retrieved in the feces of 3 of 17 mice after 7 days of supplementation and in the fecal samples of 12 of 17 mice at the end of the treatment period (Table 2). However, viable L. crispatus MU5 cells were identified only at day 14 in 2 of 14 fecal samples from the treated animals. The adherence of the probiotic strains to the colonic mucosa was evaluated at day 14, when the mice were killed; and as shown in Table 2, L. crispatus M247 was identified in 9 of 17 colonic tissue specimens, whereas we were not able to retrieve L. crispatus MU5 in any colonic tissue specimen.
TABLE 2.
Recovery of viable L. crispatus M247 and MU5 from mouse feces and tissues
| Sample type and no. of detections per mouse |
L. crispatus M247
|
L. crispatus MU5
|
||
|---|---|---|---|---|
| No. of mice whose feces contained viable probiotic strain | Mean log10 CFU/g (wet wt) of sample | No. of mice whose feces contained viable probiotic strain | Mean log10 CFU/g (wet wt) of sample | |
| Feces | ||||
| Twicea | 3 | 7.30 ± 0.44 | 0 | NDb |
| Oncec | 9 | 8.22 ± 0.25 | 2 | 8.67 ± 0.05 |
| Never | 5 | ND | 12 | ND |
| Colonic tissued | ||||
| Oncec | 9 | 7.09 ± 0.21 | 5 | 7.24 ± 0.64 |
| Never | 8 | ND | 9 | ND |
| Total no. of mice | 17 | 14 | ||
Strains M247 and MU5 were detected at days 7 and 14.
ND, not determined.
Strains M247 and MU5 were retrieved only at day 14.
Only one colonic tissue sample, obtained when the mice were killed at day 14, was analyzed.
Recovery of L. crispatus DNA from fecal samples.
Following supplementation of the diet with L. crispatus M247 and MU5, the colonic microbiota of the BALB/c mice was analyzed by PCR-DGGE. DNAs extracted from fecal samples before and after probiotic supplementation were amplified and analyzed by PCR with universal primers S-D-Bact-0968-A-S-GC and S-D-Bact-1401-a-A-17 (32) in order to amplify the V6 to V8 region of the 16S RNA gene. However, the probiotic strains administered were not retrieved in the DGGE profiles obtained with these primers (data not shown).
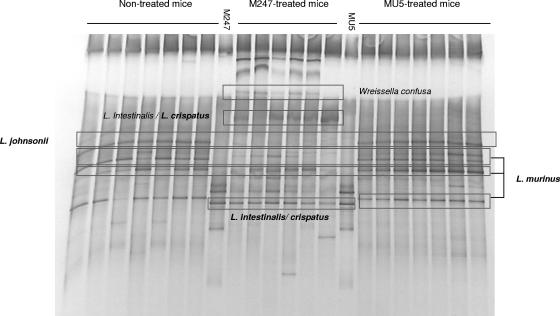
Mice receiving M247 and MU5 were therefore studied by using the DGGE primers designed by Konstantinov et al. (31, 32) and Knarreborg et al. (29) to monitor the Lactobacillus-specific bacterial community in the gastrointestinal tract. The profiles obtained for the fecal samples collected at day 0 and those collected at day 14 were compared. Several DNA fragments were excised from the gel, and their sequences were found to correspond to those of the Lactobacillus genus, as shown in Fig. 1. Indeed, L. murinus (100% identity; GenBank accession no. AF157049), as well as L. johnsonii (100% identity; GenBank accession no. AE017198), was commonly detected in the microbiota of both nontreated and probiotic-treated mice. Moreover, M247-treated mice revealed DGGE bands identified as L. intestinalis/L. crispatus (98% identity; GenBank accession no. AM117143) because of the high degree of similarity in the 16S rRNA gene sequences of these two species. However, these fragments were not retrieved in the profiles of MU5-treated mice.
FIG. 1.
Oral supplementation with aggregating strain L. crispatus M247 influences colonic microbiota. Mice received orally 108 CFU of L. crispatus M247, aggregation-deficient mutant MU5, or GG solution (vehicle) in a total volume of 100 μl for 14 days. Then, total DNA was extracted from fecal samples and amplified by nested PCR with species-specific primer pairs to identify Lactobacillus spp. The amplicons were then analyzed by PCR-DGGE. The fragments were then excised from the gel, and the eluted DNA was sequenced.
L. crispatus supplementation modulates the mRNA level of Th1 and Th2 cytokines in the colonic mucosa.
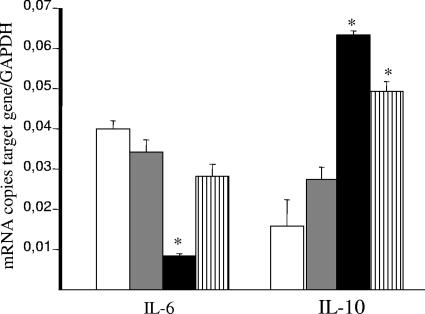
Since we recently reported that spontaneously aggregating strain L. crispatus M247 but not isogenic nonaggregating mutant MU5 reduced the severity of dextran sodium sulfate colitis in mice (13), we evaluated the effect of L. crispatus M247 and MU5 supplementation on the proinflammatory and the anti-inflammatory cytokine levels in the colonic mucosa. Thus, total RNA was extracted from the mucosa of mice supplemented for 2 weeks with L. crispatus M247 or MU5, and the amounts of the mRNAs coding for IL-6 and IL-10 were estimated by real-time quantitative RT-PCR. As shown in Fig. 2, the level of IL-6 mRNA in the colonic mucosa was significantly reduced after 2 weeks of L. crispatus M247 supplementation, whereas the level of IL-10 mRNA was significantly increased compared to that in the controls. Indeed, as shown in Fig. 2, supplementation of the diet with L. crispatus MU5 did not significantly modify the mucosal level of steady-state IL-6 and IL-10 mRNAs.
FIG. 2.
Oral supplementation with L. crispatus modifies cytokine mRNA levels in the colonic mucosa. Mice received orally 108 CFU L. crispatus M247 (black bars), aggregation-deficient mutant MU5 (gray bars), MU5 in a 30% sucrose solution that was able to reestablish the aggregation phenotype (bars with vertical stripes), or GG solution (vehicle; open bars) in a total volume of 100 μl for 14 days. Total RNA was extracted from the mucosa, and steady-state IL-6 and IL-10 mRNA levels were determined by quantitative RT-PCR and are expressed as the numbers of copies of the target gene/number of copies of the GAPDH gene (used as an internal standard) in 3 μg total RNA. For each condition, from six to eight determinations were performed, and the values are expressed as means ± SEs. *, P < 0.01 versus the results for the respective control.
L. crispatus M247 but not aggregation-deficient mutant MU5 modulates TLR2 and TLR4 levels in the colonic mucosa.
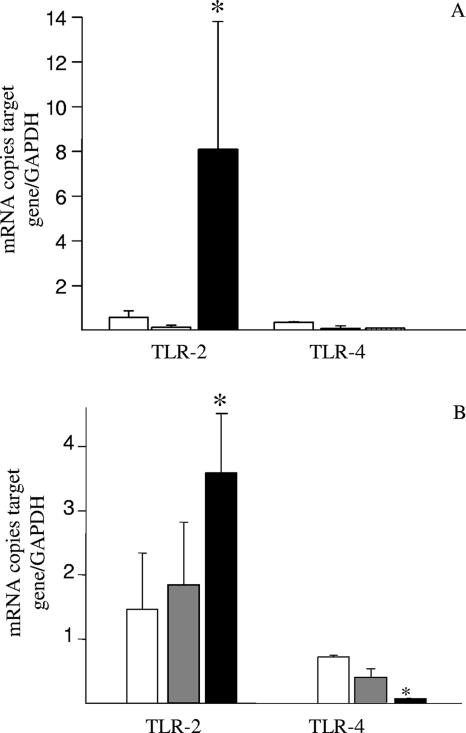
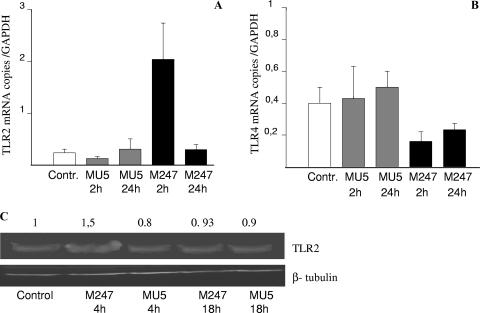
Since conserved bacterial structures modulate the activity of mucosal immune system through pattern recognition receptors such as TLRs, we decided to assess the effect of L. crispatus supplementation on the TLR2 and TLR4 levels in the colonic mucosa and in epithelial cells. As shown in Fig. 3, supplementation of the diet with L. crispatus M247 significantly increased the TLR2 mRNA levels both in the colonic mucosa and in epithelial cells, whereas the TLR4 mRNA levels were reduced in epithelial cells. These effects were evident in the colonic mucosa (Fig. 3A), as well in purified colonic epithelial cells (Fig. 3B). However, oral supplementation with the aggregation-deficient strain L. crispatus MU5 had no effect on either TLR2 and TLR4 mRNA levels (Fig. 3).
FIG. 3.
Supplementation of the diet with L. crispatus modifies TLR mRNA levels in the colonic mucosa. Mice received 108 CFU L. crispatus M247 (black bars), aggregation-deficient mutant MU5 (gray bars), or vehicle only (open bars) for 2 weeks. cDNA was prepared from total RNA extracted from the colonic mucosa (A) or from colonic epithelial cells (B), and the steady-state TLR2 and TLR4 mRNA levels were determined by quantitative RT-PCR. Values were expressed as the number of copies of TLR2 and TLR4 mRNA and were normalized to the number of copies of GAPDH mRNA (the endogenous RNA control) in 3 μg total RNA. For each condition, from six to eight determinations were performed, and values are expressed as means ± SEs. *, P < 0.01 versus the results for the control animals.
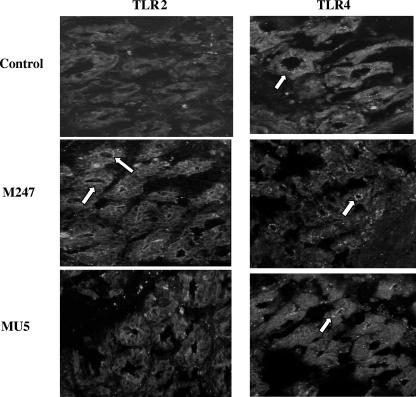
We next determined whether L. crispatus supplementation modified TLR4 and TLR2 expression and/or their distribution in the colonic mucosa. As expected, the levels of TLR4- and TLR2-specific immunostaining in the colonic mucosa of the mice were low and were localized mainly in the epithelium (Fig. 4). Indeed, following 2 weeks of oral supplementation with L. crispatus M247, a striking increase in TLR2 immunoreactivity was evident, but this was not the case with L. crispatus MU5. TLR2 staining was mainly localized in the epithelium, whereas we did not observe a significant change in mononuclear cell staining within the lamina propria. However, the intensity and distribution of TLR4-specific immunostaining were substantially unaffected in the mucosa.
FIG. 4.
Oral supplementation with L. crispatus modulates TLR4 and TLR2 protein levels in the colonic mucosa. Immunofluorescence analysis was performed with frozen sections (thickness, 10 μm) of large intestine tissue specimens collected from control mice and animals supplemented with either L. crispatus M247 or L. crispatus MU5 for 14 days. The sections were fixed in cold acetone and were incubated with an anti-TLR4 monoclonal antibody or an anti-TLR2 monoclonal antibody. Specific immunocomplexes were detected by using fluorescein isothiocyanate-labeled secondary antibodies and were visualized on a Leica TCS-NT/SP2 confocal microscope with a ×63 objective. The images are representative of four separate experiments. The arrows indicate specific immunostaining.
L. crispatus M247 directly modulates TLR expression in CMT-93 cells.
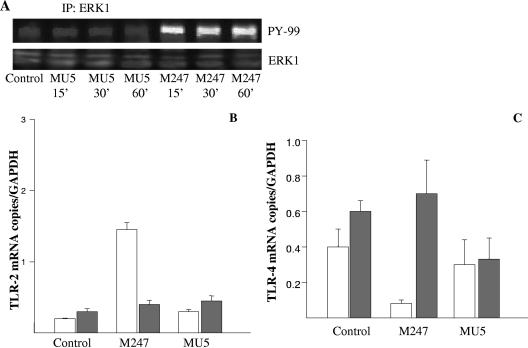
Since oral supplementation with L. crispatus not only modified the colonic microbiota but also was able to modulate cytokines and TLR expression in the colonic mucosa, we next determined whether L. crispatus directly influenced TLR levels in intestinal epithelial cells. As shown in Fig. 5, following 1 h of coculture with L. crispatus M247, CMT-93 cells showed time-dependent increases in TLR2 mRNA levels in association with a decrease in TLR4 mRNA levels, whereas aggregation-deficient mutant MU5 had no significant effects on TLR2 and TLR4 mRNA levels. As shown in Fig. 5C, Western blot analysis demonstrated a similar time-dependent increase in the TLR-2 level in CMT-93 cells exposed to L. crispatus M247 but not in cells exposed to MU5. Indeed, incubation of CMT-93 cells with 108 CFU/ml enteropathogenic E. coli did not cause any significant change in the levels of TLR2 and TLR4 expression (data not shown).
FIG. 5.
L. crispatus M247 but not L. crispatus MU5 modulates TLR4 and TLR2 levels in CMT-93 cells in vitro. CMT-93 cell monolayers were exposed to L. crispatus M247 (black bars) or MU5 (gray bars) or to medium only as a control (open bars). After 1 h of culture the medium was removed and the cells were washed and incubated for additional 1 to 24 h in fresh complete medium. (A and B) The cells were collected, the total RNA was extracted, steady-state TLR2 and TLR4 mRNA levels were determined by quantitative RT-PCR, and the values were normalized to those for GAPDH mRNA (the endogenous RNA control) in 3 μg total RNA. For each condition, nine determinations (three experiments with triplicate determinations) were performed, and the values are expressed as means ± SEs. *, P < 0.01 versus the results for control and MU5-treated cells. (C) The cells were removed, proteins soluble in RIPA buffer were extracted, and the TLR2 level was determined by Western blotting analysis. A representative blot of three different experiments with similar results is shown. The relative densitometric units of the band, with the density of the control band arbitrarily set at 1.0, are reported. The results presented here are for one of three experiments, with similar results obtained in each experiment. Beta-tubulin was used as an internal standard to confirm equal loading.
Functional relevance of L. crispatus M247-induced TLR modulation in CMT-93 cells.
Recent studies reported the ability of bacterium-derived products to modulate several activities in intestinal epithelial cells, including epithelial cell migration over the wound edge and cytokine release (15). Since we observed a striking change in TLR2 and TLR4 expression in the colonic mucosa of mice as well as in CMT-93 cells following exposure to L. crispatus M247, we assessed the functional relevance of this effect.
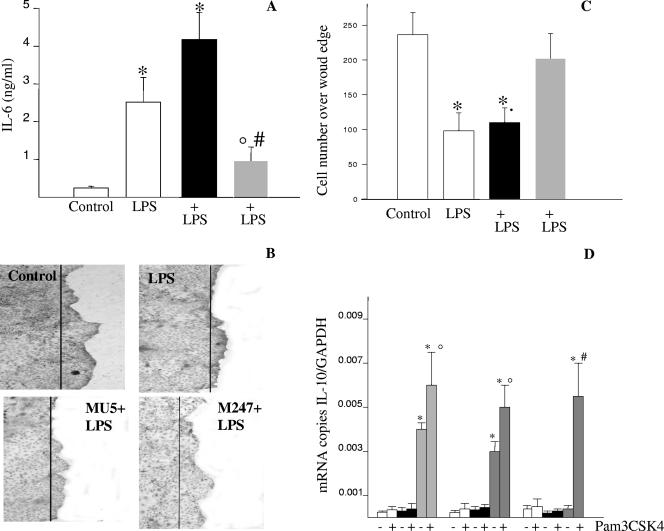
First, we determined the consequence of L. crispatus exposure on LPS-induced proinflammatory cytokine release from CMT-93 cell monolayers (Fig. 6A). As expected, LPS induced a significant release of IL-6 from CMT-93 cell monolayers. However, monolayers preincubated with L. crispatus M247, but not with aggregation-deficient mutant MU5, showed a blunted LPS-induced IL-6 release.
FIG. 6.
L. crispatus M247 but not L. crispatus MU5 modulates LPS-mediated IL-6 release and cell migration across the wound edge. CMT-93 cell monolayers were exposed to L. crispatus M247 (gray bars) or MU5 (black bars) or to medium only (open bars) as a control. After 1 h the culture medium was removed and the cells were washed and incubated in fresh complete medium. (A) Cells were incubated for an additional 6 h in medium alone or in medium containing LPS (10 μg/ml). Then, the medium was collected and the IL-6 concentration (expressed as ng/ml) was measured by enzyme-linked immunosorbent assay. For each condition, nine determinations (three experiments with triplicate determinations) were performed, and the values are expressed as means ± SEs. *, P < 0.01 versus the results for the control cells; °, P < 0.05 versus the results for the control cells; #, P < 0.01 versus the results for the cells exposed only to LPS. (B and C) Linear wounds were made with a sterile razor blade, and the cells were then incubated at 37°C in the presence of medium alone or medium supplemented with LPS (10 μg/ml). After 24 h the cells were fixed, and the nuclei were stained with hematoxylin and visualized on a Leica DM-LB inverted microscope by using a ×5 objective connected to a Leica DC-100 camera. (B) Images from a typical experiment of CMT-93 cell migration are shown, in which the line indicates the position of the cells at the wound edge. (C) Quantification of the migration rate, calculated as the mean number of epithelial cells over the wound edge, expressed as means ± SEs of three separate experiments, with 10 wound areas used to quantitate cell migration across the wound edge. (D) Cells were then incubated in medium alone or medium containing Pam3CSK4 (20 μg/ml). After 8 to 24 h, the cells were collected, total RNA was extracted, and the IL-10 mRNA level was determined by quantitative RT-PCR and expressed as the number of copies of the target gene/ the number of copies of the GAPDH gene (which was used as an internal standard) in 3 μg total RNA. For each condition, from six to eight determinations were performed, and the values are expressed as the means ± SEs. *, P < 0.01 versus the results for the respective control.
Second, we assessed the healing properties of L. crispatus treatment of CMT-93 cell monolayers in the presence of high concentrations of LPS. As expected, CMT-93 cell migration over a wound edge was significantly inhibited in the presence of LPS (Fig. 6B and C). However, in CMT-93 cell monolayers preexposed to L. crispatus M247 but not to aggregation-deficient mutant MU5, the LPS-mediated effects were abolished and significant cell migration over the wound edge was observed.
Finally, we determined the effect of Pam3CSK4, a TLR2-specific ligand, on the IL-10 mRNA level in CMT-93 cells preexposed to L. crispatus M247 or MU5. As shown in Fig. 6D, preincubation of the CMT-93 cells with L. crispatus M247, but not MU5, caused a significant increase in the IL-10 mRNA levels in the epithelial cells for up to 12 h. However, following exposure of the CMT-93 cells to L. crispatus M247, incubation with the TLR2-specific ligand resulted in a further increase in the IL-10 mRNA level that was still evident after 24 h.
The L. crispatus M247-induced TLR modulation in CMT-93 cells involves ERK activity.
Since recent studies suggested that nonpathogenic bacteria can activate specific intracellular signal cascade pathways (49, 50), we investigated the role of ERK1 in L. crispatus-induced TLR modulation. As shown in Fig. 7, incubation of CMT-93 cell monolayers with L. crispatus M247 induced a strong and time-dependent ERK1 tyrosine phosphorylation. Interestingly, aggregation-deficient mutant L. crispatus MU5, which was unable to induce significant effects on TLR mRNA and protein levels (Fig. 3 and 4), did not induce significant changes in the ERK1 tyrosine phosphorylation level. To investigate the functional relevance of ERK1 phosphorylation in M247-induced TLR modulation, we treated the CMT-93 cell monolayers with the specific ERK1 inhibitor PD98059. As depicted in Fig. 7, the inhibition of ERK1 activity significantly inhibited the effects of L. crispatus M247 on the TLR2 and TLR4 mRNAs levels in CMT-93 cell monolayers.
FIG. 7.
L. crispatus modulates TLR levels through a MAPK-dependent pathway. (A) Confluent CMT-93 cell monolayers were exposed to L. crispatus M247 or MU5 (108 CFU/ml) or to medium alone as a control for 15 to 60 min, and then the medium was removed and the cells were washed and lysed by addition of RIPA buffer. The cell lysates (2 mg/ml) were then incubated with a rabbit anti-ERK1 polyclonal antibody (2 h at 4°C), and the immunoprecipitated proteins were then fractionated on an SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. Phosphorylated ERK1 was detected by using anti-phosphotyrosine pY99. A representative blot of three different experiments with similar results obtained in each experiment is shown. (B and C) Confluent CMT-93 cell monolayers were incubated for 30 min in the presence of the ERK1 inhibitor PD98059 (10 μM) (gray bars) or medium alone (open bars). Then, the monolayers were exposed to L. crispatus M247 or MU5 or to medium only as a control. After 1 h of culture the medium was removed and the cells were washed and incubated for additional 1 h in fresh complete medium. The cells were then collected, the total RNA was extracted, the steady-state TLR2 and TLR4 mRNA levels were determined by quantitative RT-PCR, and the values were normalized to the mRNA level of GAPDH, which acted as an endogenous RNA control. For each condition, nine determinations (three experiments with triplicate determinations) were performed, and the values are expressed as the means ± SEs. *, P < 0.01 versus the results for the control and the MU5-treated cells.
DISCUSSION
An imbalance in the endogenous intestinal microflora is now considered a critical component in the chain of events contributing to the development of dysfunctional immune responses by the host's mucosa-associated immune system, leading to the onset of many clinically relevant diseases (60). Indeed, several clinical trials that used live bacteria (i.e., probiotics) to manipulate the intestinal flora for the treatment of acute and chronic diseases have obtained encouraging results (8, 54). However, the choice of biotherapeutic agents is still based on empirical approaches, and comprehensive knowledge of the bacteria's relevant phenotypic traits necessary to induce beneficial effects on the host's microbiota and mucosal immune system is lacking. In this study we identified a phenotypic characteristic associated with a probiotic strain required to guarantee its persistence in the gastrointestinal tract, to shape the intestinal microflora, and to modulate in intestinal epithelial cells the expression of TLRs, a class of receptors able to deeply affect the activities of the innate and adaptive mucosal immune responses following microbe recognition.
Probiotic candidates for therapeutic applications are generally screened by using heterogeneous models on the basis of their ability to survive in the presence of gastric acid, tolerate bile salts, and adhere to gut mucus and epithelial cell monolayers in vitro but by paying no attention to the identification of the bacterial characteristics required to exert the beneficial effects in vivo (35, 53). Therefore, it is not surprising that the probiotic bacteria chosen by this strategy often fail to exert the desired biological effect (21). Here we report that the aggregation phenotype, often observed within Lactobacillus spp., is associated with a stronger ability to colonize the intestine and to produce immunomodulatory effects in vitro and in vivo. Indeed, in a previous study we reported that the aggregation phenotype was associated with protective effects in a colitis model in vivo (13). The ability to aggregate or coaggregate has been demonstrated in several bacterial species colonizing harsh environments, such as the oral cavity and the intestinal mucosa (38, 48). Indeed, several Lactobacillus spp. show a strong aggregating phenotype and the property of coaggregation with E. coli strains and enterococci (14, 30). This phenotypic property may provide greater chances for survival, persistence, and colonization of the host's mucosal surfaces and, therefore, to come into strict contact with the mucosal immune system.
The indigenous commensal microflora that initiates innate immune responses plays an active role in host mechanisms that maintain tissue homeostasis (16, 26, 39). Since commensal bacteria differ in their ability to promote the development and activity of gut-associated lymphoid tissue, changes in the colonic ecosystem may have profound effects on the host's health (61). In this scenario, membrane and cytoplasmic receptors in intestinal immune and nonimmune cells play a key role recognizing repetitive patterns of nonpathogenic gram-positive and gram-negative microbes since PRR-derived signals influence a variety of physiological activities. In fact, TLR-induced signaling regulates the synthesis of cytoprotective factors (47) essential for intestinal barrier function and repair (47) and stimulates the release of antimicrobial peptides and immunomodulatory cytokines (40, 44, 59). However, excessive TLR stimulation can have deleterious effects on the host (i.e., it can trigger persistent inflammation); therefore, TLR signaling is carefully regulated in the healthy gut by several mechanisms, including the anatomical distribution of receptors, as well as the level distribution of accessory and regulatory molecules (41). Thus, the down-regulation of TLR cell surface expression and the inhibition of intracellular signaling might contribute to the tolerance to normal bacterial products observed in intestinal epithelial cells (42). Conversely, abnormal TLR expression/signaling has been associated with inflammatory bowel diseases (1, 12) and increased sensitivity to the development of colitis in mice (46, 55). As a matter of fact, in this study we report that following oral supplementation with L. crispatus M247, the TLR mRNA and protein levels in the intestinal mucosa and epithelial cells were profoundly modified, since the level of TLR4 was drastically reduced, whereas TLR2 was up-regulated. Indeed, this may represent an additional mechanism involved in the regulation of the intestinal microbiota of the host's mucosal immune system, since regulation of the level of expression of a receptor determines the sensitivity of a system to a biological stimulus (22). Therefore, probiotics like L. crispatus M247 and L. casei, which increase the level of mucosal TLR2 expression over that of TLR4, might establish a higher level of mucosal sensitivity to commensal nonpathogenic gram-positive bacteria, such as lactic acid bacteria, bifidi, and Enterococcus spp., generally considered to exert favorable effects on the mucosal immune system function (27).
Although TLR signaling is required to promote tissue homeostasis, the role of different receptors may be quite different in this regard. Thus, TLR2-derived signaling mainly promotes Th2-type cytokine release, whereas TLR4 activation by LPS primarily stimulates Th1-type responses (2). In addition, TLR2 stimulation induces dendritic cell maturation (45) and protection from pathogens through the secretion of antimicrobial peptides (37) and enhances the mucosal barrier function by up-regulating the expression of ZO1 (10). On the other end, the down-regulation of TLR4 expression protects intestinal epithelial cells from the deleterious responses triggered by gram-negative commensal bacteria by inducing the release of an excess of proinflammatory cytokines and inhibits epithelial cell migration over a wound edge (1, 46). Therefore, it is not surprising that in a healthy intestine the level of TLR4 expression in intestinal epithelial cells is low and that the increased level of expression that occurs in patients with inflammatory bowel disease is associated with the loss of tolerance toward commensal bacteria (12). In addition, the modulation of TLR expression might result in the observed immunomodulatory effects on mucosal cytokine levels associated with probiotic administration that lead to increased IL-10 levels as opposed to reduced IL-1 levels (18). In fact, we observed that the undesired effects of TLR4 stimulation, such as IL-6 release and inhibition of epithelial cell migration over the wound edge, and the beneficial consequences of TLR2 stimulation, like IL-10 expression, in intestinal epithelial cells are profoundly affected by L. crispatus M247 exposure as a consequence of TLR modulation, thus suggesting that at least part of the beneficial effects of lactobacilli are mediated through the fine-tuning of the TLR expression profile in intestinal epithelial cells. Indeed, a paper by Cario et al. published while this paper was under revision demonstrated that the administration of a TLR2 synthetic ligand is effective at reducing the inflammatory damage in mice, further supporting the relevance of TLR2 modulation following probiotic administration (11).
Intestinal epithelial cells release a number of factors affecting the mucosa-associated microbiota: mucus, defensins, and enzymes which contribute to shape the colonic flora (23). However, bacteria profoundly influence intestinal epithelial cell and mucosa-associated lymphoid tissue function by directly transferring to the epithelial cells or releasing a variety of molecules in the extracellular environment (23, 51). Thus, bacterium-derived products (i.e., LPS, muramyl dipeptide, lipoteichoic acid, and esotoxins) bind to specific receptors and trigger different intracellular signal transduction cascades, leading to phenotypic modifications (62) or to the release of an array of soluble mediators, such as cytokines and prostaglandins. Thus, the typology of the soluble mediators released is strikingly different, depending on the bacteria, either pathogens or commensal anti-inflammatory bacteria, from which they arise (25). The physiological significance of the distinct modes of action of pathogenic and anti-inflammatory gut bacteria is not fully appreciated, since they seem to share signal transduction components, such as the NF-κB signaling pathway, which is also involved in a variety of functions, such as cellular integrity, survival, and repair (4, 20, 28). However, recent studies suggest that nonpathogenic bacteria may activate specific intracellular signal cascades. Thus, probiotic bacteria such as Lactobacillus rhamnosus GG and Bacteroides lactis induce NF-κB activation and p38 mitogen-activated protein kinase (MAPK) signaling cascades, whereas Bacteroides vulgatus, a commensal able to trigger colitis in genetically predisposed hosts, triggers NF-κB activation but not p38 MAPK phosphorylation (50). Here, we show that aggregation-deficient strain L. crispatus MU5, which is devoid of protective and immunomodulatory efficacy in vivo and in vitro, as opposed to aggregating strain M247, was unable to activate the ERK1 signaling pathway, supporting a key role for MAPK signaling in the epithelium in response to Lactobacillus strains showing probiotic activity. Furthermore, a recent study by Resta-Lenert and Barrett has reported that probiotic-mediated protection of the epithelial cell damage produced by inflammatory cytokines also requires MAPK signaling, underscoring the key role of this signal pathway in the effects of probiotic bacteria on intestinal epithelial cells (49).
In conclusion, we identified a phenotypic trait for a probiotic strain, Lactobacillus crispatus, that is associated with the ability to persist and colonize the host's colon as well as to significantly modify the colonic microbiota. Aggregation-competent strain L. crispatus M247, which is able to exert favorable effects on a model of intestinal inflammation in vivo (13), significantly modifies the expression in colonic epithelial cells of TLR4 and TLR2, two key receptors in the innate immune system, following in vivo or in vitro exposure. Therefore, probiotic strains able to affect the mucosal expression of key receptors for bacterial conserved structures can establish the responsiveness of the mucosa-associated immune system to the commensal flora and therefore modulate the feedback mechanisms which regulate mucosal immune responses to the constant challenge by luminal bacteria (62).
Acknowledgments
This study was supported in part by grant 2004063577_004 from MIUR to G.C.S.
Footnotes
Published ahead of print on 18 July 2007.
REFERENCES
- 1.Abreu, M. T., P. Vora, E. Faure, L. S. Thomas, E. T. Arnold, and M. Arditi. 2001. Decreased expression of Toll-like receptor-4 and MD-2 correlates with intestinal epithelial cell protection against dysregulated proinflammatory gene expression in response to bacterial lipopolysaccharide. J. Immunol. 1671609-1616. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, S., A. Agrawal, B. Doughty, A. Gerwitz, J. Blenis, T. Van Dyke, and B. Pulendran. 2003. Cutting edge: different Toll-like receptors agonist instruct dendritic cells to induce distinct Th responses via different modulation of extracellular signal-regulated kinase-mitogen-activated protein kinase and c-Fos. J. Immunol. 1714984-4989. [DOI] [PubMed] [Google Scholar]
- 3.Amies, C. S. 1967. A modified formula for the preparation of Stuart's transport medium. Can. J. Public Health 58296-300. [PubMed] [Google Scholar]
- 4.Bambou, J. C., A. Giraud, S. Menard, B. Begue, S. Rakotobe, M. Heyman, F. Taddei, N. Cerf-Bensussan, and V. Gaboriau-Routhiau. 2004. In vitro and ex vivo activation of the TLR5 signaling pathway in intestinal epithelial cells by a commensal Escherichia coli strain. J. Biol. Chem. 27942984-42992. [DOI] [PubMed] [Google Scholar]
- 5.Beutler, B. 2004. Inferences, questions and possibilities in Toll-like receptor signaling. Nature 430257-263. [DOI] [PubMed] [Google Scholar]
- 6.Bibiloni, R., M. A. Simon, C. Albright, B. Sartor, and G. W. Tannock. 2005. Analysis of the large bowel microbiota of colitic mice using PCR/DGGE. Lett. Appl. Microbiol. 4145-51. [DOI] [PubMed] [Google Scholar]
- 7.Blum, S., and E. J. Schiffrin. 2003. Intestinal microflora and homeostasis of the mucosal immune response: implications for probiotic bacteria? Curr. Issues Intest. Microbiol. 453-60. [PubMed] [Google Scholar]
- 8.Broekaert, I. J., and W. A. Walker. 2006. Probiotics and chronic disease. J. Clin. Gastroenterol. 40270-274. [DOI] [PubMed] [Google Scholar]
- 9.Cario, E. 2005. Bacterial interactions with cells of the intestinal mucosa: Toll-like receptors and NOD2. Gut 541182-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cario, E., G. Gerken, and D. Podolsky. 2004. Toll-like receptor 2 enhances ZO-1-associated intestinal epithelial barrier integrity via protein kinase C. Gastroenterology 127224-238. [DOI] [PubMed] [Google Scholar]
- 11.Cario, E., G. Gerken, and D. K. Podolsky. 2007. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology 1321359-1374. [DOI] [PubMed] [Google Scholar]
- 12.Cario, E., and D. K. Podolsky. 2000. Differential alteration in intestinal epithelial cell expression of Toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect. Immun. 687010-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castagliuolo, I., F. Galeazzi, S. Ferrari, M. Elli, P. Brun, A. Cavaggioni, D. Tormen, G. C. Sturniolo, L. Morelli, and G. Palù. 2005. Beneficial effect of auto-aggregating Lactobacillus crispatus on experimentally induced colitis in mice. FEMS Immunol. Med. Microbiol. 43197-204. [DOI] [PubMed] [Google Scholar]
- 14.Cesena, C., L. Morelli, M. Alander, T. Siljander, E. Tuomola, S. Salminen, T. Mattila-Sandholm, and A. von Wright. 2001. Lactobacillus crispatus and its non aggregating mutant in human colonization trials. J. Dairy Sci. 841001-1010. [DOI] [PubMed] [Google Scholar]
- 15.Cetin, S., H. R. Ford, L. R. Sysko, C. A. Wang, M. D. Neal, C. Baty, G. Apodaca, and D. J. Hackam. 2004. Endotoxin inhibits intestinal epithelial restitution through activation of rho-GTPase and increased focal adhesions. J. Biol. Chem. 27924592-24600. [DOI] [PubMed] [Google Scholar]
- 16.Coyne, M. J., B. Reinap, M. M. Lee, and L. E. Comstock. 2005. Human symbionts use a host-like pathway for surface fucosylation. Science 3071778-1781. [DOI] [PubMed] [Google Scholar]
- 17.Deplancke, B., K. Finster, W. V. Graham, C. T. Collier, J. E. Thurmond, and H. R. Gaskins. 2003. Gastrointestinal and microbial responses to sulfate-supplemented drinking water in mice. Exp. Biol. Med. 228424-433. [DOI] [PubMed] [Google Scholar]
- 18.Dieleman, L. A., M. S. Goerres, A. Arends, D. Sprengers, C. Torrice, F. Hoentjen, W. B. Grenther, and R. B. Sartor. 2003. Lactobacillus GG prevents recurrence of colitis in HLA-B27 transgenic rats after antibiotic treatment. Gut 52370-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galdeano, C. M., and G. Perdigon. 2006. The probiotic bacterium Lactobacillus casei induces activation of the gut immune system through innate immunity. Clin. Vaccine Immunol. 13219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galdiero, M., M. Vitiello, E. Sanzari, M. D'Isanto, A. Tortora, A. Longanella, and S. Galdiero. 2002. Porins from Salmonella enterica serovar Typhimurium activate the transcription factors activating protein 1 and NF-κB through the Raf-1-mitogen-activated protein kinase cascade. Infect. Immun. 70558-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudier, E., C. Michel, C. Cherbut, and C. Hoebler. 2005. The VSL#3 probiotic mixture modifies microflora but does not heal chronic dextran-sodium sulfate-induced colitis or reinforce the mucus barrier in mice. J. Nutr. 1352753-2761. [DOI] [PubMed] [Google Scholar]
- 22.Harper, P. A., D. S. Riddick, and A. B. Okey. 2006. Regulating the regulator: factors that control levels and activity of the aryl hydrocarbon receptor. Biochem. Pharmacol. 72267-279. [DOI] [PubMed] [Google Scholar]
- 23.Hect, G. 1999. Innate mechanisms of epithelial host defense: spotlight on intestine. Am. J. Physiol. 277(3 Pt 1)C351-C358. [DOI] [PubMed] [Google Scholar]
- 24.Heilig, H., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagnoff, M. F., and L. Eckmann. 2001. Analysis of host responses to microbial infection using gene expression profiling. Curr. Opin. Microbiol. 4246-250. [DOI] [PubMed] [Google Scholar]
- 26.Kelly, D., J. I. Campbell, T. P. King, G. Grant, E. A. Jansson, A. G. Coutts, S. Pettersson, and S. Conway. 2004. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat. Immunol. 5104-112. [DOI] [PubMed] [Google Scholar]
- 27.Kelly, D., S. Conway, and R. Aminov. 2005. Commensal gut bacteria: mechanisms of immune modulation. Trends Immunol. 26326-333. [DOI] [PubMed] [Google Scholar]
- 28.Kim, Y. G., T. Ohta, T. Takahashi, A. Kushiro, K. Nomoto, T. Yokokura, N. Okada, and H. Danbara. 2006. Probiotic Lactobacillus casei activates innate immunity via NF-κB and p38 MAP kinase signaling pathways. Microbes Infect. 8994-1005. [DOI] [PubMed] [Google Scholar]
- 29.Knarreborg, A., M. A. Simon, R. C. Engberg, B. B. Jensen, and G. W. Tannock. 2002. Effect of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens of various ages. Appl. Environ. Microbiol. 685918-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolebrander, P. E. 1991. Co-aggregation: adherence in the human oral microbial ecosystem, p. 303-309. In M. Dworkin (ed.), Microbial cell-cell interactions. American Society for Microbiology, Washington, DC.
- 31.Konstantinov, S. R., A. Awati, H. Smidt, B. A. Williams, A. D. L. Akkermans, and W. M. de Vos. 2004. Specific response of a novel and abundant Lactobacillus amylovorus-like phylotype to dietary prebiotics in the guts of weaning piglets. Appl. Environ. Microbiol. 703821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konstantinov, S. R., W.-Y. Zhu, B. A. Williams, S. Tamminga, and W. M. de Vos. 2003. Effect of fermentable carbohydrates on faecal bacterial communities as revealed by DGGE analysis of 16S rDNA. FEMS Microbiol. Ecol. 43225-235. [DOI] [PubMed] [Google Scholar]
- 33.Lan, J. C., S. M. Cruickshank, J. C. Singh, M. Farrar, J. P. Lodge, P. J. Felsburg, and S. R. Carding. 2005. Different cytokine response of primary colonic epithelial cells to commensal bacteria. World J. Gastroenterol. 113375-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lotz, M. M., I. Rabinovitz, and A. M. Mercurio. 2000. Intestinal restitution: progression of actin cytoskeleton rearrangements and integrin function in a model of epithelial wound healing. Am. J. Pathol. 156985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mao, Y., S. Nobaek, B. Kasravi, D. Adawi, U. Stenram, G. Molin, and G. Jeppson. 1996. The effect of Lactobacillus strains and oat fibre on methotrexate-induced enterocolitis in rats. Gastroenterology 111334-344. [DOI] [PubMed] [Google Scholar]
- 36.McCraken, V. J., J. M. Simpson, R. I. Macie, and H. R. Gaskins. 2001. Molecular ecological analysis of dietary and antibiotic-induced alterations of the mouse intestinal microbiota. J. Nutr. 1311862-1870. [DOI] [PubMed] [Google Scholar]
- 37.Melmed, G., L. S. Thomas, N. Lee, S. Y. Tesfay, K. Kukasek, K. S. Michelsen, Y. Zhou, B. Hu, M. Arditi, and M. T. Abreu. 2003. Human intestinal epithelial cells are broadly unresponsive to Toll-like receptor 2-dependent bacterial ligands: implication for host-microbial interaction in the gut. J. Immunol. 1701406-1415. [DOI] [PubMed] [Google Scholar]
- 38.Millsap, K. W., H. C. van der Mei, R. Bos, and H. J. Busscher. 1998. Adhesive interactions between medically important yeasts and bacteria. FEMS Microbiol. Rev. 21321-336. [DOI] [PubMed] [Google Scholar]
- 39.Neish, A. S., A. T. Gewirtz, H. Zeng, A. N. Young, M. E. Hobert, V. Karmali, A. S. Rao, and J. L. Madara. 2000. Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alpha ubiquitination. Science 2891560-1563. [DOI] [PubMed] [Google Scholar]
- 40.Netea, M. G., J. W. Van der Meer, and B. J. Kullberg. 2004. Toll-like receptors as an escape mechanism from the host defense. Trends Microbiol. 12484-488. [DOI] [PubMed] [Google Scholar]
- 41.Ortega-Cava, C. F., S. Ishihara, M. A. K. Rumi, K. Kawashima, N. Ishimura, H. Kazumori, J. Udagawa, Y. Kadowaki, and Y. Kinishita. 2003. Strategic compartmentalization of Toll-like receptor 4 in the mouse gut. J. Immunol. 1703977-3985. [DOI] [PubMed] [Google Scholar]
- 42.Otte, J. M., E. Cario, and K. Podolsky. 2004. Mechanism of cross hyporesponsiveness to toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology 1261054-1070. [DOI] [PubMed] [Google Scholar]
- 43.Reference deleted.
- 44.Pasare, C., and R. Medzhitov. 2003. Toll pathway-dependent blockade of CD4+CD25+ T cell-mediated suppression by dendritic cells. Science 2991033-1036. [DOI] [PubMed] [Google Scholar]
- 45.Pulendran, B. 2005. Modulating vaccine response with dendritic cells and Toll-like receptors. J. Immunol. 12457-2465. [DOI] [PubMed] [Google Scholar]
- 46.Qureshi, F. G., C. Leaphart, S. Cetin, J. Li., A. Grishin, S. Watkins, H. R. Ford, and D. J. Hackam. 2005. Increased expression and function of integrins in enterocytes by endotoxin impairs epithelial restitution. Gastroenterology 1281012-1022. [DOI] [PubMed] [Google Scholar]
- 47.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzihitov. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118229-241. [DOI] [PubMed] [Google Scholar]
- 48.Reniero, R., P. Cocconcelli, V. Bottazzi, and L. Morelli. 1992. High frequency of conjugation in Lactobacillus mediated by an aggregation-promoting factor. J. Gen. Microbiol. 138763-768. [Google Scholar]
- 49.Resta-Lenert, S., and K. E. Barrett. 2006. Probiotics and commensals reverse TNF-alpha- and IFN-gamma-induced dysfunction in human intestinal epithelial cells. Gastroenterology 130731-746. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz, P. A., M. Hoffmann, S. Szcesny, M. Blaut, and D. Haller. 2005. Innate mechanisms for Bifidobacterium lactis to activate transient pro-inflammatory host responses in intestinal epithelial cells after the colonization of germ-free rats. Immunology 115441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruppoli, R., and M. G. Pizza. 2000. Bacterial toxins, p. 193-280. In P. Cossard, P. Boquet, S. Normark, and G. Ruppoli (ed.), Cellular microbiology. American Society for Pharmacology and Biology, Washington, DC.
- 52.Salzman, N. H., H. de Jong, Y. Paterson, H. J. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 1483651-3660. [DOI] [PubMed] [Google Scholar]
- 53.Sartor, R. B. 1997. Enteric microflora in IBD: pathogens or commensal? Inflamm. Bowel Dis. 3230-235. [PubMed] [Google Scholar]
- 54.Sazawal, S., G. Hiremath, U. Dhingra, P. Malik, S. Deb, and R. Black. Efficacy of probiotics in prevention of acute diarrhoea: a meta-analysis of masked, randomised, placebo-controlled trials. Lancet Infect. Dis. 6374-382. [DOI] [PubMed]
- 55.Sebastiani, G., G. Laveque, L. Lariviere, L. Laroche, E. Skamene, and P. Gros. 2000. Cloning and characterization of the murine Toll-like receptor (Tlr5) gene: sequence and mRNA expression studies in salmonella susceptible MOLF/Ei mice. Genomics 64230-240. [DOI] [PubMed] [Google Scholar]
- 56.Shen, L., and J. R. Turner. 2006. Role of epithelial cells in initiation and propagation of intestinal inflammation. Eliminating the stating tight junction dynamics exposed. Am. J. Physiol. Gastrointest. Liver Physiol. 290G577-G582. [DOI] [PubMed] [Google Scholar]
- 57.Reference deleted.
- 58.Reference deleted.
- 59.Thoma-Uszynski, S., S. Stenger, O. Takeuchi, M. Engele, P. A. Sieling, P. F. Barnes, M. Rollinghoff, P. L. Bolcskei, M. Wagner, S. Akira, M. V. Norgard, J. T. Belisle, P. J. Godwoski, B. R. Bloom, and R. L. Modlin. 2001. Induction of direct antimicrobial activity through mammalian Toll-like receptors. Science 2911544-1547. [DOI] [PubMed] [Google Scholar]
- 60.Tlaskalova-Hogenova, H., R. Stepankova, T. Hudcovic, L. Tuckova, B. Cukrowska, R. Lodinova-Zadnikova, H. Kozakova, P. Rossmann, J. Bartova, D. Sokol, D. P. Funda, D. Borovska, Z. Rehakova, J. Sinkora, J. Hofman, P. Drastich, and A. Kokesova. 2004. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol. Lett. 9397-108. [DOI] [PubMed] [Google Scholar]
- 61.Umesaki, Y., H. Setoyama, S. Matsumoto, A. Imoaka, and K. Itoh. 1999. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 673504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uzzau, S., and A. Fasano. 2000. Cross-talk between enteric pathogens and the intestine. Cell. Microbiol. 283-89. [DOI] [PubMed] [Google Scholar]
- 63.Vaathvuo, J., P. Toivanen, and E. Eerola. 2001. Study of murine faecal microflora by cellular fatty acid analysis; effect of age and mouse strain. Antonie Leeuwenhoek 8035-42. [DOI] [PubMed] [Google Scholar]
- 64.Zoetendal, E. G., A. D. L. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoretic analysis of 16S rRNA from fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 643854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]