Abstract
Cross-reactivity between Mycobacterium kansasii ESAT-6 and CFP-10 homologues and their M. bovis counterparts can confound the interpretation of immunodiagnostic tests for tuberculosis. M. kansasii is a nontuberculous mycobacterial species cultured from skin test-positive cattle in Great Britain. Using peptides derived from M. bovis and M. kansasii ESAT-6 and CFP-10 regions that differ between these species, we investigated the species specificity and cross-reactivity at the level of individual bovine T-cell epitopes. Our results demonstrated that all peptides tested are fully cross-reactive, with the exception of one ESAT-6-derived peptide that harbored an M. bovis-specific epitope(s) when it was recognized in the context of bovine leukocyte antigen (BoLA)-DQ but that was cross-reactive with its M. kansasii homologues when it was restricted by BoLA-DR. This observation further highlights that prediction of species specificity by comparing sequence identity/homology alone is not sufficient and that individuals with diverse major histocompatibility complex constellations need to be tested to characterize the cross-reactivity or species specificity of peptide-based reagents.
The incidence of bovine tuberculosis (BTB) in Great Britain has been increasing steadily over the last two decades (10). At present the BTB control program involves regular skin testing with the single intradermal comparative tuberculin test, followed by the compulsory slaughter of cattle with a positive result (19). Tuberculin specificity can also be affected by infection of cattle by nontuberculous mycobacterial (NTM) species, as can be seen by the frequent background responses to avian tuberculin that is being used to control for such environmental sensitization. Encouragingly, when antigens like ESAT-6 and CFP-10 are used in the BOVIGAM gamma interferon (IFN-γ) test, they have also been shown to enhance the specificity of the tuberculin-based test per se in this respect (7, 8, 20, 21, 26, 31). However, some NTM species, like Mycobacterium kansasii, M. gordonae, M. marinum, M. szulgai, M. flavescens, and M. gastri, contain genes for ESAT-6 and CFP-10 homologues (4, 20). Cattle infected with such ESAT-6- or CFP-10-expressing NTM species could give rise to false-positive responses to these antigens. This has been demonstrated in a recent study by Waters et al. (32), who demonstrated potent cross-reactive IFN-γ responses with the M. bovis ESAT-6 and CFP-10 protein species in cattle experimentally infected with M. kansasii. Genes for M. kansasii homologues have also been described for other M. tuberculosis antigens, such as MPB83, TB10.4, and TB10.3 (24). M. kansasii can cause disease in cattle presenting with lesions in the pulmonary tract and associated lymph nodes (18). This organism can also cause pulmonary disease in immunocompetent humans and disseminated disease almost exclusively in immunocompromised individuals (3, 12). In vitro responses to M. tuberculosis ESAT-6 and CFP-10 in humans infected with M. kansasii have been reported, again suggesting cross-reactivity between the M. tuberculosis and M. kansasii protein homologues. Interestingly, M. kansasii is highly heterogenic, with five subtypes/genotypes being described (2). Subtype I has been found exclusively in humans; in contrast, subtype II can be isolated from both human and environmental samples, whereas subtypes III to V are rarely found in humans but were present in environmental samples (2, 4). The amino acid sequences of the ESAT-6 and CFP-10 homologues of these M. kansasii genotypes have been defined and were found to be highly homologous with those of M. tuberculosis (6). Armed with this sequence information, we decided to investigate the potential species specificity and cross-reactivity between the M. bovis and M. kansasii ESAT-6 and CFP-10 homologues at the molecular level, i.e., at the level of the individual peptide epitopes recognized by CD4+ T cells isolated from cattle infected with M. bovis. We also investigated with this synthetic peptide-based approach whether M. bovis-specific reagents could be defined.
MATERIALS AND METHODS
Antigens and peptides.
Bovine tuberculin (purified protein derivative [PPD] B) and avian tuberculin (PPD A) were supplied by the Tuberculin Production Unit at the Veterinary Laboratories Agency (VLA), Weybridge, Surrey, United Kingdom, and were used in vitro in IFN-γ enzyme-linked immunospot (ELISPOT) assays or enzyme immunoassays (EIAs) at 10 μg/ml. Staphylococcal enterotoxin B was used as a positive control at 1 μg/ml. Synthetic peptides derived from the sequences of M. bovis and M. kansasii ESAT-6 and CFP-10 (see Table 1 for the sequences) were synthesized by the facility of the University of Leiden (Leiden, The Netherlands) by solid-phase synthesis and purified, and their quality was assessed as described previously (24). The peptides were used at 25 μg/ml in the IFN-γ ELISPOT assay. Recombinant M. tuberculosis ESAT-6 and CFP-10 were kindly provided by M. Singh (Gesellschaft für Biotechnologische Forschung, Braunschweig, Germany) and were used at 5 μg/ml in in vitro assays (IFN-γ EIA and ELISPOT assay).
TABLE 1.
Peptides used in this study
| Protein and peptide | Peptide sequencea | Species (Mka genotype)b |
|---|---|---|
| ESAT-6 | ||
| 1 | EAYQGVQQKWDATATE | M. bovis |
| 1A | --------------Q- | M. kansasii (I-V) |
| 2 | KWDATATELNNALQNL | M. bovis |
| 2A | ------Q--------- | M. kansasii (I-IV) |
| 2B | ------Q---S----- | M. kansasii (V) |
| 3 | LNNALQNLARTISEAG | M. bovis |
| 3A | --------S------- | M. kansasii (I-IV) |
| 3B | --S-----S------- | M. kansasii (V) |
| CFP-10 | ||
| 4 | QEAGNFERISGDLKTQ | M. bovis |
| 4A | ------------M--- | M. kansasii (IV) |
| 5 | IDQVESTAGSLQGQWR | M. bovis |
| 5A | --------A---A--- | M. kansasii (I-V) |
| 6 | GSLQGQWRGAAGTAAQAA | M. bovis |
| 6A | A---A------------- | M. kansasii (I-V) |
| 6B | A---A------A------ | M. kansasii (IV) |
| 7 | AGTAAQAAVVRFQEAANK | M. bovis |
| 7A | --A--------------- | M. kansasii (IV) |
| 8 | VVRFQEAANKQKQELDEI | M. bovis |
| 8A | ------------A--E-- | M. kansasii (I-V) |
| 9 | QKQELDEISTNIRQAGVQYS | M. bovis |
| 9A | --A----E--------------- | M. kansasii (I-V) |
| 10 | NIRQAGVQYSRADEEQQQ | M. bovis |
| 10A | ----------K------- | M. kansasii (I-V) |
-, amino acid residue in M. kansasii-derived peptide identical to the residue in the M. bovis sequence. Amino acids are represented by the one-letter code.
M. kansasii genotype according to Arend et al. (4).
Cattle used in this study.
Heparinized blood samples were obtained from adult tuberculin skin test-positive cattle (n = 15) from herds with ongoing BTB cases. Seven animals were selected due to the presence of strong IFN-γ responses after stimulation with recombinant ESAT-6 and/or CFP-10; IFN-γ was evaluated with the BOVIGAM EIA kit (see below). These cows were of five different breeds (Holstein-Friesian, Charolais, Devon, British Friesian, and Aberdeen Angus) and were obtained from five herds from distinct geographical locations within Great Britain. The disease status of the animals was confirmed postmortem by the presence of lesions in lymph nodes and/or lungs typical of BTB and by the culture of M. bovis from tissues. The animals were held at VLA under the provision of a Home Office project license granted under the Animals (Scientific Procedures) Act, 1986. This license and the experiments listed in it were approved by the local ethical review committee.
Whole-blood IFN-γ assay (BOVIGAM assay) (33).
To select animals for further study, blood samples were collected from M. bovis-infected cattle and were placed into heparinized Vacutainer tubes (Becton Dickinson, Oxford, United Kingdom). Briefly, whole-blood cultures were performed in 96-well tissue culture plates (flat bottom) in the presence of tuberculin and recombinant ESAT-6 and CFP-10. After 24 h of culture at 37°C, the plasma supernatants (100 μl/well) were harvested and stored at −20°C. The BOVIGAM IFN-γ ELISA was performed according to the supplier's instructions (Prionics, Schlieren, Switzerland). Animals showing recombinant ESAT-6- or CFP-10-specific changes in the values of the optical density at 450 nm of >0.5 were selected for this study.
Ex vivo IFN-γ ELISPOT assay (28).
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood by Histopaque-1077 (Sigma) gradient centrifugation and were immediately cultured in RPMI 1640 tissue culture medium (Life Technologies, Paisley, Scotland, United Kingdom) supplemented with 10% fetal calf serum (Sigma Aldrich, Poole, United Kingdom), nonessential amino acids (Sigma Aldrich), 5 × 10−5 M 2-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate. The ex vivo (direct) ELISPOT assay results were enumerated as described earlier (28). Briefly, ELISPOT assay plates (Immunobilon-P polyvinylidene difluoride membranes; Millipore, Molsheim, France) were coated overnight at 4°C with the bovine IFN-γ-specific monoclonal antibody 5D10 (BioSource, Wheatley, United Kingdom). Unbound antibody was removed by washing, and the wells were blocked with 10% fetal calf serum in RPMI 1640 medium. PBMCs suspended in tissue culture medium were then added (2 × 105 PBMCs/well) and cultured in the presence of PPD B, recombinant proteins, or synthetic peptides for 24 h at 37°C in a humidified 5% CO2 incubator. Spots were developed with rabbit serum specific for IFN-γ prepared at VLA, followed by incubation with an alkaline phosphatase-conjugated monoclonal antibody specific for rabbit immunoglobulin G (Sigma Aldrich). The spot-forming cells (SFCs) were visualized with 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrate (Sigma Aldrich). The SFCs in the wells with medium only were subtracted from the SFCs in wells with antigen or peptides (ΔSFCs/well). ΔSFCs/well of >10 were considered positive. For blockage of the bovine leukocyte antigen (BoLA) presentation of peptides, monoclonal antibodies specific for BoLA-DR and BoLA-DQ were purchased from VRMD, Pullman, WA. Each antibody was preincubated with PBMCs in the ELISPOT assay plate at 5 μg/ml for 1 h at 37°C, after which the relevant peptide was added to the wells and the plates were incubated as described above for the standard IFN-γ ELISPOT assays (13).
Hain genotype system.
Forty-six frozen nontuberculous isolates from the Weybridge TB Section culture collection were used. Once they were thawed, 300 μl of each isolate was sown onto one Lowenstein-Jensen-pyruvate medium slope and one Lowenstein-Jensen-glycerol medium slope, and the slopes were incubated at 37°C. Once the isolate had grown sufficiently on the medium, 1 μl was harvested into 100 μl of high-pressure liquid chromatography-grade water and heat killed at 80°C for half an hour. The resulting heat-killed isolate was then used to perform PCR analysis. The Hain genotype kit (Hain Lifescience, Nehren, Germany) (22) was used to test the samples, according to the supplier's instructions. A positive result was obtained when the staining reaction of the kit produced readable bands on the test strip, and the results were interpreted by using the supplier's template.
RESULTS AND DISCUSSION
NTM in cattle in Great Britain.
Only a small percentage of mycobacterial isolates obtained from the tissues of cattle slaughtered under the test and slaughter control policy in Great Britain are NTM (ca. 0.8% of total isolates from 2004 to 2006). The majority of these NTM isolates belong to the M. avium/M. intracellulare (MAI) complex (for example, 54% of the NTM isolates in Great Britain from 2004 to 2006). The remaining NTM isolates remained unclassified or comprised a number of different mycobacterial species. Recently, the species of 46 non-MAI NTM isolates (5 from cows with visible lesions typical of BTB and 41 from cattle presenting without visible lesions postmortem) were determined by using the Hain genotype system (22). The results indicated that 22/46 of the non-MAI NTM isolates tested were M. kansasii, including isolates from two of five of the lesioned animals. Genotyping revealed that all M. kansasii isolates found in these cattle were subtype I or II, which could be isolated from humans in a study in Holland (4). The other species found were M. celatum (5/46); M. nonchromogenicum (5/46); M. gordonae (5/46); M. szulgai (4/46); M. fortuitum (2/46); and M. scrofulaceum, M. intermedium, and M. shimodei (1/46 each). These results do not constitute the findings of a representative study of NTM isolates in Great Britain. They do demonstrate, however, that M. kansasii can be isolated from cattle tissues. As stated above, apart from M. kansasii, M. gordonae and M. szulgai were also isolated, and these species also contain genes for ESAT-6 homologues (4) and most likely also genes for CFP-10 homologues. Therefore, these three NTM species can potentially express these cross-reactive antigens. M. kansasii isolates have also been identified in an analysis of NTM cattle isolates from Northern Ireland, although the dominant NTM species were part of the MAI complex (46%) and the M. terrae complex (39%), with 8% of the isolates being M. kansasii (20).
These data highlighted the potential need for assessment of the cross-reactivities of the M. kansasii and M. bovis homologues of ESAT-6 and CFP-10, although as referred to above, the actual percentage of NTM species in general and M. kansasii in particular compared to that of M. bovis among isolates from cattle is low. However, an additional observation is that M. kansasii can also be isolated from humans without disease: one-third of isolates have been reported to represent colonization rather than infection (1), although it is not known if this could lead to T-cell responses. Colonization in the absence of disease also occurs in cattle (20) and could therefore comprise a possible source of false-positive responses to tuberculin and antigens like ESAT-6 and CFP-10, with resulting greater potential financial and epidemiological impacts beyond those from the relatively few clinical cases. Due to the potential geographical tropism of M. kansasii infections (2), such “false-positive” responders are unlikely to be equally distributed throughout Great Britain and could therefore be a problem of particular herds. Indeed, herds with consistent and protracted cases of animals that are skin test reactors, that do not have lesions typical of BTB, and from whose tissues M. bovis cannot be cultured have been observed. The reasons for these so-called nonspecific skin test reactions are not known, and there is yet no evidence that they are due to M. kansasii infection. Nevertheless, it would be desirable to be able to develop immunodiagnostic reagents that can distinguish between M. bovis- and M. kansasii-infected cattle.
Comparison of sequence homologies between M. bovis/M. tuberculosis and M. kansasii CFP-10 and ESAT-6 homologues in relation to the positions of dominant promiscuous bovine T-cell epitopes.
The sequences of the M. kansasii homologue genes of ESAT-6 and CFP-10 of all five M. kansasii genotypes and the deduced amino acid sequences have been reported previously (4). The M. bovis and M. tuberculosis CFP-10 and ESAT-6 molecules are identical (9, 14) and are henceforth referred to in this paper as M. bovis homologues. Comparison of the M. kansasii homologues with the respective M. bovis sequences demonstrated a high degree of identity (>95%) at the amino acid level (4). For example, the ESAT-6 homologues differ only at two positions (M. kansasii subtypes I to IV) or three positions (M. kansasii subtype V) (Fig. 1), whereas the CFP-10 homologues differed between the M. bovis protein and the different M. kansasii subtypes by five to seven residues (Fig. 1). As previous studies have shown, ESAT-6- and CFP-10-derived epitopes are recognized promiscuously by class II-restricted T cells from tuberculous cattle (27), and it is therefore possible to assess the potential impact of the high degree of sequence identity between the M. kansasii and M. bovis homologues in relation to these immunodominant epitopes. The epitopes within the sequences of CFP-10 and ESAT-6 that were recognized by CD4+ T cells from >50% of the cattle tested are shown in Fig. 1 (27, 29, 31; H. M. Vordermeier et al., unpublished data). The amino acid sequences of the majority of these epitopic regions are fully identical between the protein homologues (i.e., the N-terminal epitopes from both ESAT-6 and CFP-10). Full cross-recognition of these sequences after M. bovis or M. kansasii infection is therefore almost inevitable. In contrast, other dominant epitopes differed at between one and two amino acid residues between the M. bovis- and M. kansasii-derived proteins (e.g., within ESAT-6 peptides 49 to 64) (Fig. 1), and these differences could be sufficient to impart species specificity to such peptides. The unambiguous prediction of T-cell cross-reactivity is difficult even in situations with high degrees of sequence identity between epitope regions. For example, while we could demonstrate that a high degree of sequence identity between sequence regions from unrelated mycobacterial antigens (>50% in the 16- to 20-mer regions) indicated cross-reactivity at the epitope level in cattle, other peptides that displayed similar degrees of sequence identity were not cross-reactive (17). Conversely, the molecular basis of T-cell cross-reactivity between two unrelated mycobacterial proteins can depend on the sequence identity of 1 residue of 8 to 10 residues spanning the epitope (16). Moreover, changing one single residue within a 20-mer amino acid peptide containing a T-cell epitope can result in the peptide derivative not being recognized by T cells specific for the wild-type peptide (30), e.g., by abrogating major histocompatibility complex (MHC) binding. To highlight this point, we performed single-alanine mutation analysis of ESAT-6 peptides 49 to 64. While single substitutions of most residues with alanine did not affect or only marginally affected T-cell recognition (data not shown), replacement of T63 with alanine resulted in a complete loss of T-cell responses (data not shown). Interestingly, T63 in the M. bovis sequence has been replaced by a glutamine residue in the M. kansasii homologues (4). Therefore, despite the high degrees of sequence homology between the M. bovis and M. kansasii homologues of CFP-10 and ESAT-6, it seemed of interest to investigate the potential species specificity and cross-reactivity at the level of individual peptides recognized by CD4+ T cells isolated from cattle infected with M. bovis and to determine whether a synthetic peptide-based approach could define “M. bovis-specific” peptides useful for the differentiation between M. kansasii and M. bovis infections.
FIG. 1.
Alignment of the amino acid sequences and positions of dominant T-cell epitopes of ESAT-6 (A) and CFP-10 (B) of M. bovis/M. tuberculosis and M. kansasii genotypes I to V (MKA-1 to MKA-V). Amino acids are represented by the one-letter code. Promiscuous recognition of M. bovis-derived ESAT-6 and CFP-10 peptides (defined as giving rise to T-cell responses in ≥50 of the cattle tested that recognized M. bovis ESAT-6 or CFP-10) is represented by open bars above the sequences (27, 31). The proportions of animals (responder frequencies) that recognized these peptides are shown above the respective bars (27, 31). (Adapted from reference 4 with permission of the publisher.)
Recognition of M. bovis- and M. kansasii-derived ESAT-6 and CFP-10 peptides by T cells from cattle with BTB.
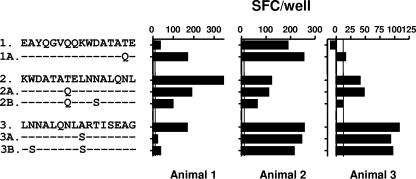
Synthetic peptides representing the regions of M. kansasii ESAT-6 and CFP-10 with sequence differences compared to the sequences of their M. bovis counterparts were prepared (five and eight peptides, respectively; Table 1). The recognition of these peptides was compared to the recognition of the M. bovis-derived peptides (Table 1): PBMCs from tuberculous cattle known to recognize either CFP-10 or ESAT-6 (or both) were prepared and stimulated with these peptides to determine the IFN-γ responses by ELISPOT assay. The results of these experiments are summarized in Fig. 2 (ESAT-6) and Fig. 3 (CFP-10). With the exception of M. bovis CFP-10-derived peptide 6, all 10 M. bovis-derived peptides were recognized by at least one cow, thereby demonstrating the presence of epitopes (Fig. 2 and 3). The results for ESAT-6-derived peptides are shown in Fig. 2. Peptides 1 and 1A and peptides 2, 2A, and 2B, encompassing two of the three sequence regions of ESAT-6 with differences between M. bovis and M. kansasii, were recognized by at least one animal tested, thereby confirming immunological cross-reactivity at the epitope level. The most interesting results were obtained with peptides 3, 3A, and 3B (representing sequences derived from M. bovis, M. kansasii subtypes I to IV, and M. kansasii subtype V, respectively) (Fig. 2). In animal 3 only M. bovis-derived peptide 3 was recognized, as shown in Fig. 2 and 4C, thus suggesting species-specific responses to this peptide. In contrast, however, the majority of cattle tested (three of five cattle tested; see the results for animals 2 and 3 in Fig. 2 and Fig. 4A) recognized peptides 3, 3A, and 3B equally well; i.e., they displayed fully cross-reactive responses.
FIG. 2.
Recognition of M. bovis and M. kansasii ESAT-6-derived peptides by T cells from cattle with BTB. PBMCs from naturally infected cattle were stimulated with synthetic peptides (see Table 1 for sequence details). The readout system indicates the IFN-γ production measured by the ex vivo ELISPOT assay. The results are expressed as the mean SFCs/well, and the standard errors of the means for all values are <20% of the means. Horizontal lines indicate the cutoff for positivity (10 ΔSFCs/well).
FIG. 3.
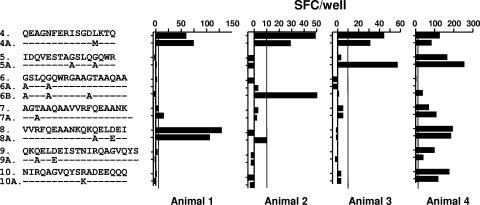
Recognition of M. bovis and M. kansasii CFP-10-derived peptides by T cells from cattle with BTB. PBMCs from naturally infected cattle were stimulated with synthetic peptides (see Table 1 for sequence details). The readout system indicates the IFN-γ production measured by the ex vivo ELISPOT assay. The results are expressed as the mean SFCs/well, and the standard errors of the means for all values are <20% of the means. Horizontal lines indicate the cutoff for positivity (10 ΔSFCs/well).
FIG. 4.
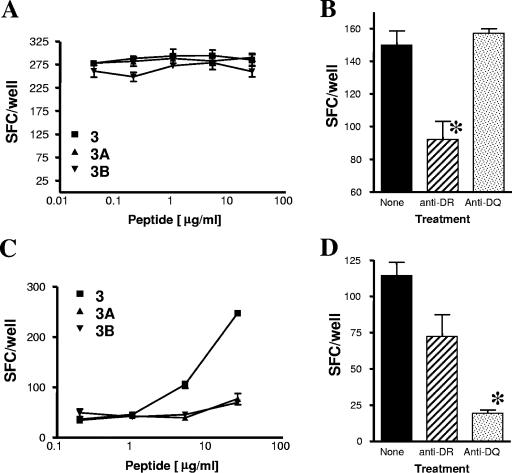
BoLA class II restriction of the recognition of M. bovis ESAT-6 peptide 3. Cross-reactive recognition of peptides 3, 3A, and 3B in animal 2 (A) was BoLA-DR restricted (B). M. bovis-specific recognition of peptide 3 in animal 1 (C) was BoLA-DQ restricted (D). Anti-BoLA-DR or anti-BoLA-DQ monoclonal antibody was added to peptide 3-stimulated PBMC cultures (B and D). The results are expressed as the mean numbers of IFN-γ-producing SFCs/well ± standard errors of the means. *, P < 0.05 compared to the results for the other groups (Student's t test).
The M. kansasii CFP-10-derived peptides (peptides 4A to 10A) were all recognized by at least one tuberculous cow tested, thus demonstrating full cross-reactivity between M. bovis and M. kansasii CFP-10 peptide homologues (Fig. 3). Interestingly, in animal 2 M. kansasii-derived peptide 6B (representing subtype IV) was recognized, whereas M. bovis-derived peptide 6 was not (Fig. 3). A similar observation was made in respect to the recognition of peptides 5 and 5A by T cells from animal 3; peptide 5A was strongly recognized, while the corresponding M. bovis peptide was not (although both peptide species were recognized in animal 4) (Fig. 3).
It has been shown that promiscuously recognized peptides can harbor different sets of epitopes that can be recognized in the context of different MHC class II alleles (e.g., for peptides from a mycobacterial antigen) (30), thus accounting for their promiscuous recognition across a range of different MHC class II alleles. We therefore hypothesized that the “M. bovis specificity” of ESAT-6 peptide 3 in some animals, in contrast to the cross-reactivity with peptides 3A and 3B in others, may be due to the recognition of distinct epitopes within peptide 3 that are recognized in the context of different BoLA class II types or alleles. Thus, we next defined the restriction elements used in ESAT-6 peptide 3 recognition by blocking the IFN-γ responses with monoclonal antibodies specific for BoLA-DR and BoLA-DQ. Two animals were selected: animal 2, which recognized peptides 3, 3A, and 3B in a cross-reactive manner (Fig. 4A), and animal 1, which recognized only peptide 3 (Fig. 4C). Interestingly, recognition of peptide 3 was BoLA-DR restricted in the animal that recognized both the M. bovis- and the M. kansasii-derived peptides (Fig. 4B), whereas the “species-specific” recognition of peptide 3 in animal 1 was restricted by BoLA-DQ (Fig. 4D). Similar results were obtained with a second similar pair of cattle tested (data not shown). The BoLA complex consists of one DR gene pair and up to two DQ gene pairs per haplotype and is highly polymorphic (11, 15). We had previously shown that identical peptides from mycobacterial antigens can be recognized in cattle in the context of both BoLA-DR and BoLA-DQ molecules (13), as had been reported previously, for example, for the antigen part of a vaccine against foot-and-mouth disease virus (25). Our results therefore confirm and extend these earlier observations. In addition, our data on cross-reactivity when peptide 3 was recognized in the context of BoLA-DR or when “M. bovis specificity” was recognized in the context of BoLA-DQ suggest that this is because the 20-mer peptide harbors distinct epitope regions that bind differentially to BoLA-DR or BoLA-DQ. Interestingly, peptides that are derived from mycobacterial antigens such as CFP-10 and that are recognized by human CD4+ cells in the context of HLA-DR and HLA-DQ have also been described (5, 23).
In summary, this study has extended studies by Waters et al. (32) by assessing the T-cell cross-reactivity between the M. bovis/M. tuberculosis and the M. kansasii homologues of ESAT-6 and CFP-10 at the peptide/epitope level by testing synthetic peptides derived from both mycobacterial species. Generally, we found, with the possible exception of ESAT-6 peptide 6, complete cross-reactivity between the homologue peptides in infected cattle. It is therefore unlikely that one can design peptide-based reagents that would allow the immunological differentiation of cattle infected with M. bovis or M. kansasii even if further antigens like MPB83 (34) and TB10.4 were to be considered. Furthermore, we could show that identical peptides can be recognized in the context of BoLA-DR and BoLA-DQ and that the restriction element used determined whether such peptides are “M. bovis specific” or cross-reactive. This observation introduces a further complication in attempts to predict species specificity by comparing sequence identity/homology alone and highlights that empirical experimental studies with individuals that express diverse BoLA class II haplotypes are required to assess T-cell cross-reactivity.
Acknowledgments
This study was funded by the Department for Environment, Food and Rural Affairs, United Kingdom.
This study would not have been possible without the contribution of the State Veterinary Service, in particular, Linda Farrant, in identifying naturally M. bovis-infected, tuberculin-positive cattle. We also express our appreciation to the staff of the Animal Service Unit at VLA, in particular, Derek Clifford, for their dedication to the welfare of the cattle housed at VLA.
Footnotes
Published ahead of print on 1 August 2007.
REFERENCES
- 1.Ahn, C. H., J. W. McLarty, S. S. Ahn, S. I. Ahn, and G. A. Hurst. 1982. Diagnostic criteria for pulmonary disease caused by Mycobacterium kansasii and Mycobacterium intracellulare. Am. Rev. Respir. Dis. 125388-391. [DOI] [PubMed] [Google Scholar]
- 2.Alcaide, F., I. Richter, C. Bernasconi, B. Springer, C. Hagenau, R. Schulze-Robbecke, E. Tortoli, R. Martin, E. C. Bottger, and A. Telenti. 1997. Heterogeneity and clonality among isolates of Mycobacterium kansasii: implications for epidemiological and pathogenicity studies. J. Clin. Microbiol. 351959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arend, S. M., E. Cerda de Palou, P. de Haas, R. Janssen, M. A. Hoeve, E. M. Verhard, T. H. Ottenhoff, D. van Soolingen, and J. T. van Dissel. 2004. Pneumonia caused by Mycobacterium kansasii in a series of patients without recognised immune defect. Clin. Microbiol. Infect. 10738-748. [DOI] [PubMed] [Google Scholar]
- 4.Arend, S. M., P. de Haas, E. Leyten, I. Rosenkrands, L. Rigouts, P. Andersen, W. Mijs, J. T. van Dissel, and D. van Soolingen. 2005. ESAT-6 and CFP-10 in clinical versus environmental isolates of Mycobacterium kansasii. J. Infect. Dis. 1911301-1310. [DOI] [PubMed] [Google Scholar]
- 5.Arend, S. M., A. Geluk, K. E. van Meijgaarden, J. T. van Dissel, M. Theisen, P. Andersen, and T. H. Ottenhoff. 2000. Antigenic equivalence of human T-cell responses to Mycobacterium tuberculosis-specific RD1-encoded protein antigens ESAT-6 and culture filtrate protein 10 and to mixtures of synthetic peptides. Infect. Immun. 683314-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arend, S. M., K. E. van Meijgaarden, K. de Boer, E. C. de Palou, D. van Soolingen, T. H. Ottenhoff, and J. T. van Dissel. 2002. Tuberculin skin testing and in vitro T cell responses to ESAT-6 and culture filtrate protein 10 after infection with Mycobacterium marinum or M. kansasii. J. Infect. Dis. 1861797-1807. [DOI] [PubMed] [Google Scholar]
- 7.Buddle, B. M., A. R. McCarthy, T. J. Ryan, J. M. Pollock, H. M. Vordermeier, R. G. Hewinson, P. Andersen, and G. W. de Lisle. 2003. Use of mycobacterial peptides and recombinant proteins for the diagnosis of bovine tuberculosis in skin test-positive cattle. Vet. Rec. 153615-620. [DOI] [PubMed] [Google Scholar]
- 8.Buddle, B. M., T. J. Ryan, J. M. Pollock, P. Andersen, and G. W. de Lisle. 2001. Use of ESAT-6 in the interferon-gamma test for diagnosis of bovine tuberculosis following skin testing. Vet. Microbiol. 8037-46. [DOI] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 10.De La Rua, R. 2006. Bovine tuberculosis in the European Union and other countries: current status, control programmes and constraints to eradication. Gov. Vet. J. 1619-45. [Google Scholar]
- 11.Ellis, S. A., and K. T. Ballingall. 1999. Cattle MHC: evolution in action? Immunol. Rev. 167159-168. [DOI] [PubMed] [Google Scholar]
- 12.Evans, A. J., A. J. Crisp, R. B. Hubbard, A. Colville, S. A. Evans, and I. D. Johnston. 1996. Pulmonary Mycobacterium kansasii infection: comparison of radiological appearances with pulmonary tuberculosis. Thorax 511243-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewer, K., P. Cockle, S. Gordon, H. Mansoor, M. Govaerts, K. Walravens, S. Marche, G. Hewinson, and M. Vordermeier. 2006. Antigen mining with iterative genome screens identifies novel diagnostics for the Mycobacterium tuberculosis complex. Clin. Vaccine Immunol. 1390-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 1007877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glass, E. J. 2004. Genetic variation and responses to vaccines. Anim. Health Res. Rev. 5197-208. [DOI] [PubMed] [Google Scholar]
- 16.Harris, D. P., H. M. Vordermeier, M. Singh, C. Moreno, S. Jurcevic, and J. Ivanyi. 1995. Cross-recognition by T cells of an epitope shared by two unrelated mycobacterial antigens. Eur. J. Immunol. 253173-3179. [DOI] [PubMed] [Google Scholar]
- 17.Hewinson, R. G., H. M. Vordermeier, N. H. Smith, and S. V. Gordon. 2006. Recent advances in our knowledge of Mycobacterium bovis: a feeling for the organism. Vet. Microbiol. 112127-139. [DOI] [PubMed] [Google Scholar]
- 18.Jarnagin, J. L., E. M. Himes, W. D. Richards, D. W. Luchsinger, and R. Harrington, Jr. 1983. Isolation of Mycobacterium kansasii from lymph nodes of cattle in the United States. Am. J. Vet. Res. 441853-1855. [PubMed] [Google Scholar]
- 19.Office International des Epizooties. 1959. Zoonoses: second report of the joint WHO/FAO Expert Committee. Office International des Epizooties, Paris, France.
- 20.Pollock, J. M., and P. Andersen. 1997. The potential of the ESAT-6 antigen secreted by virulent mycobacteria for specific diagnosis of tuberculosis. J. Infect. Dis. 1751251-1254. [DOI] [PubMed] [Google Scholar]
- 21.Pollock, J. M., R. M. Girvin, K. A. Lightbody, R. A. Clements, S. D. Neill, B. M. Buddle, and P. Andersen. 2000. Assessment of defined antigens for the diagnosis of bovine tuberculosis in skin test-reactor cattle. Vet. Rec. 146659-665. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz, P., J. Gutierrez, F. J. Zerolo, and M. Casal. 2002. GenoType mycobacterium assay for identification of mycobacterial species isolated from human clinical samples by using liquid medium. J. Clin. Microbiol. 403076-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shams, H., P. Klucar, S. E. Weis, A. Lalvani, P. K. Moonan, H. Safi, B. Wizel, K. Ewer, G. T. Nepom, D. M. Lewinsohn, P. Andersen, and P. F. Barnes. 2004. Characterization of a Mycobacterium tuberculosis peptide that is recognized by human CD4+ and CD8++ T cells in the context of multiple HLA alleles. J. Immunol. 1731966-1977. [DOI] [PubMed] [Google Scholar]
- 24.Skjot, R. L., I. Brock, S. M. Arend, M. E. Munk, M. Theisen, T. H. Ottenhoff, and P. Andersen. 2002. Epitope mapping of the immunodominant antigen TB10.4 and the two homologous proteins TB10.3 and TB12.9, which constitute a subfamily of the esat-6 gene family. Infect. Immun. 705446-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Lierop, M. J., P. R. Nilsson, J. P. Wagenaar, J. M. Van Noort, J. D. Campbell, E. J. Glass, I. Joosten, and E. J. Hensen. 1995. The influence of MHC polymorphism on the selection of T-cell determinants of FMDV in cattle. Immunology 8479-85. [PMC free article] [PubMed] [Google Scholar]
- 26.van Pinxteren, L. A., P. Ravn, E. M. Agger, J. Pollock, and P. Andersen. 2000. Diagnosis of tuberculosis based on the two specific antigens ESAT-6 and CFP10. Clin. Diagn. Lab. Immunol. 7155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vordermeier, M., A. O. Whelan, and R. G. Hewinson. 2003. Recognition of mycobacterial epitopes by T cells across mammalian species and use of a program that predicts human HLA-DR binding peptides to predict bovine epitopes. Infect. Immun. 711980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vordermeier, H. M., M. A. Chambers, P. J. Cockle, A. O. Whelan, J. Simmons, and R. G. Hewinson. 2002. Correlation of ESAT-6-specific gamma interferon production with pathology in cattle following Mycobacterium bovis BCG vaccination against experimental bovine tuberculosis. Infect. Immun. 703026-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vordermeier, H. M., P. C. Cockle, A. Whelan, S. Rhodes, N. Palmer, D. Bakker, and R. G. Hewinson. 1999. Development of diagnostic reagents to differentiate between Mycobacterium bovis BCG vaccination and M. bovis infection in cattle. Clin. Diagn. Lab. Immunol. 6675-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vordermeier, H. M., D. P. Harris, C. Moreno, and J. Ivanyi. 1994. Promiscuous T cell recognition of an H-2 IA-presented mycobacterial epitope. Eur. J. Immunol. 242061-2067. [DOI] [PubMed] [Google Scholar]
- 31.Vordermeier, H. M., A. Whelan, P. J. Cockle, L. Farrant, N. Palmer, and R. G. Hewinson. 2001. Use of synthetic peptides derived from the antigens ESAT-6 and CFP-10 for differential diagnosis of bovine tuberculosis in cattle. Clin. Diagn. Lab. Immunol. 8571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waters, W. R., M. V. Palmer, T. C. Thacker, J. B. Payeur, N. B. Harris, F. C. Minion, R. Greenwald, J. Esfandiari, P. Andersen, J. McNair, J. M. Pollock, and K. P. Lyashchenko. 2006. Immune responses to defined antigens of Mycobacterium bovis in cattle experimentally infected with Mycobacterium kansasii. Clin. Vaccine Immunol. 13611-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood, P. R., and S. L. Jones. 2001. BOVIGAM: an in vitro cellular diagnostic test for bovine tuberculosis. Tuberculosis (Edinburgh) 81147-155. [DOI] [PubMed] [Google Scholar]
- 34.Woolford, A. J., R. G. Hewinson, M. Woodward, and J. W. Dale. 1997. Sequence heterogeneity of an mpb70 gene analogue in Mycobacterium kansasii. FEMS Microbiol. Lett. 14843-48. [DOI] [PubMed] [Google Scholar]