During cell division, eukaryotic cells must faithfully pass on their genetic material to the next generation during mitosis. It has long been known that lower eukaryotes and higher eukaryotes achieve this in strikingly different ways. Higher eukaryotes undergo an open mitosis in which the nuclear envelope is completely disassembled at the G2/M transition and is not reassembled until after DNA segregation in telophase/G1. In contrast, many lower eukaryotes undergo a closed mitosis in which the nuclear envelope remains intact and mitosis occurs within the nucleus. However, classifying mitosis as being either open or closed has its limitations. Many early studies using phase and electron microscopy indicated that fungi have evolved variations in how the mitotic segregation of the DNA and the nuclear envelope is achieved (1, 2, 16, 38). For example, even in organisms which break down their nuclear envelope during mitosis, the phase during mitosis when the nuclear envelope breaks down can vary between organisms (16). Many of these early studies are described in an extensive and excellent review by Heath (16), which summarized the different morphological characteristics of many fungi and other lower eukaryotes during mitosis. More recently, the availability of fungal genome sequences combined with molecular genetics and live cell imaging has shed light on the mechanisms regulating such variant mitoses. Here we review recent advances in our understanding of fungal mitosis and discuss how the biology of different organisms and cell types may have necessitated variant mitoses.
The physical process of segregating duplicated chromosomes occurs on one of the most striking and dynamic of all subcellular structures, the mitotic spindle. Simplistically, the mitotic spindle is composed of microtubules which extend from each spindle pole and connect to the kinetochore region of chromosomes. Microtubules are composed of α and β tubulin subunits, and microtubule lengthening and shortening are important to segregate chromosomal DNA. One problem facing cells undergoing a closed mitosis is the need to relocalize tubulin and mitotic regulators from the cytoplasm to the nucleus in order to form a spindle within the nucleus. If the nuclear envelope is intact, the only way in and out of the nucleus is through nuclear pore complexes (NPCs) (17, 51). Therefore, in organisms undergoing a closed mitosis, tubulin must gain access to the nucleus through the NPC in order to form a spindle. NPCs are embedded in the nuclear envelope and act as molecular sieves, selectively facilitating the transport of proteins and nucleic acids in and out of the nucleus. Each nucleus contains many NPCs, which are composed of ∼30 different proteins called nucleoporins, or Nups (17). The basic overall structure of the NPC is conserved between lower and higher eukaryotes, and many of the individual Nups can be identified in highly divergent species based on sequence homology (17). Not surprisingly, some Nups contain transmembrane domains and likely anchor the NPC in the nuclear envelope. Other Nups are part of a core ring structure which transits the nuclear envelope. The center of this ring structure is termed the central channel and acts as the gateway between the nucleus and cytoplasm (52, 53). The central channel of the NPC is occupied by a class of Nups which contain phenylalanine-glycine (FG) repeats and are termed FG-repeat Nups (39, 51, 52). In addition, other Nups form cytoplasmic fibrils and a nuclear basket (51). The FG-repeat Nups restrict the diffusion of macromolecules through the NPC central channel and also have the ability to bind transport factors carrying their cargo (11, 23, 39, 52). The binding of FG-repeat Nups to cargo-ladened transport factors helps facilitate active transport through the NPC in a Ran-dependent manner (5, 39, 45, 52).
MODIFYING NUCLEAR TRANSPORT DURING CLOSED MITOSIS
In organisms undergoing an open mitosis, the NPC is disassembled along with the nuclear envelope, and therefore nuclear transport does not occur during open mitoses. However, in organisms undergoing a closed mitosis, the NPC must still function as a conduit between the nucleus and the cytoplasm. Extensive studies of the model budding yeast Saccharomyces cerevisiae indicate that the NPC remains intact throughout mitosis in this organism (4, 18, 20, 28). Therefore, tubulin and proteins which need to get into nuclei to regulate mitotic entry must do so by mitosis-specific active transport through the NPC. It is worth noting, however, that S. cerevisiae differs from many other organisms undergoing closed mitosis in that spindle formation begins during DNA replication in S phase. Therefore, a large influx of tubulin immediately prior to mitosis may not be necessary in this organism. In S. cerevisiae, the mechanisms regulating changes in nuclear transport between interphase and mitosis are at least in part controlled by mitotic inhibition of the Kap121 nuclear transport pathway (27). Interestingly, inhibition of the Kap121-mediated nuclear transport is regulated by the binding of Kap121 to a phosphorylated form of Nup53 (27), suggesting that mitotic changes in nuclear transport may be regulated at the level of the NPC. During interphase, Nup53 is bound to Nup170, and Kap121-mediated nuclear transport is active. In contrast, during mitosis, phosphorylated Nup53 binds to Kap121, which is thought to cause inhibition of Kap121-mediated nuclear transport and thus help regulate mitosis (27).
OPENING THE NUCLEAR PORE COMPLEX TO ALLOW MITOTIC ENTRY
Partial disassembly of the nuclear pore complex.
Until recently, modification of nuclear transport pathways was presumed to be the mechanism that regulates the nuclear entry of tubulin and mitotic regulators in fungi undergoing closed mitosis. However, studies of the Aspergillus nidulans mitosis-specific never-in-mitosis (NIMA) kinase have uncovered another mechanism by which proteins can gain access through the NPC during mitotic entry. This essential kinase was first identified by Ron Morris in a genetic screen aimed at identifying genes with cell cycle functions or with roles in nuclear migration (30). NIMA kinase activity is absolutely required for mitotic entry, and its inactivation causes cells to arrest in late G2, even though the Cdk1/cyclin B kinase is fully activated (34). Studies to determine how NIMA regulates mitotic entry indicate that its primary target is likely the NPC (6, 7, 55). These studies aimed to identify mutations in other genes which allowed nimA1 mutants arrested in G2 to enter mitosis. Mutations in only two supressor of nimA1 (son) genes, sonA and sonB, were isolated in this screen, although the isolation of sonA mutations multiple times suggests that this screen was near saturation (6, 55). Initial studies indicated that these genes encoded two physically interacting Nups, SONBnNup98 and SONAGle2 (Nup98 and Rae1 in humans) (6). The question then arose, how could these Nups be involved in NIMA-activated mitotic entry? The localization of SONBnNup98 and SONAGle2 during the cell cycle gave significant insight into this question. During interphase, SONBnNup98 and SONAGle2 localized to the nuclear periphery, as expected for Nups. However, during mitosis, both SONBnNup98 and SONAGle2 dramatically dispersed from the NPC and relocated throughout the cell (7). Clearly the NPC does not remain intact during A. nidulans closed mitosis, but to what extent does it disassemble? Using a systematic approach, 24 Nups were identified in the A. nidulans genome, based on sequence homology with known Nups in S. cerevisiae and humans. These were endogenously tagged with the green fluorescent protein (GFP) moiety and analyzed for cell cycle-dependent changes in their localization. Remarkably, 14 of these Nups dispersed from the NPC during mitosis while the others remained located at the nuclear periphery (7, 33). Thus, a massive reorganization of the NPC occurs specifically during mitosis. An example of the differential mitotic locations of A. nidulans Nups can be seen in the time-lapse images shown in Fig. 1A. Nup96 remains at the nuclear periphery throughout mitosis, while in contrast, Nup49 disperses from the nuclear periphery during mitosis but returns during mitotic exit.
FIG. 1.
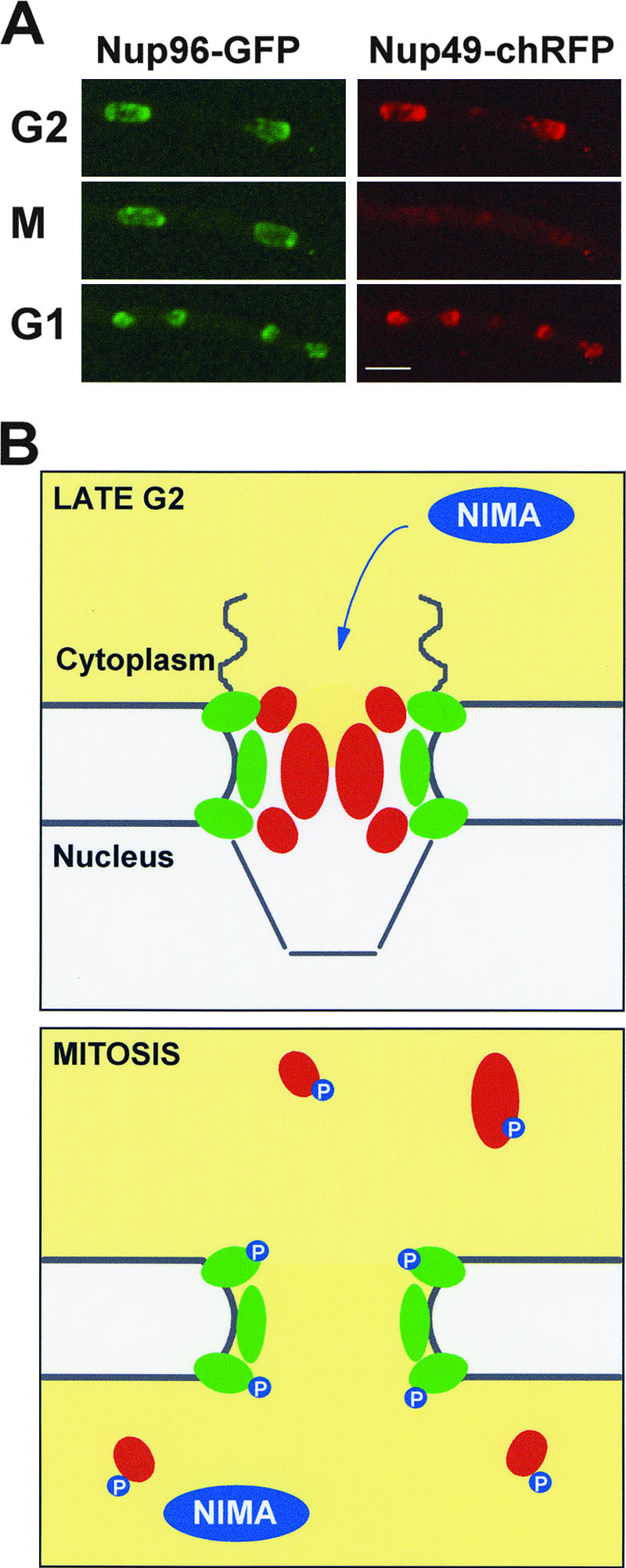
Partial NPC disassembly during mitosis in A. nidulans. (A) Time-lapse images of Nup96 labeled with GFP and Nup49 labeled with the mCherry variant of red fluorescent protein (chRFP) in the same cell (43). During G2, both Nups locate to NPCs at the nuclear periphery. However, as the cell enters mitosis, Nup49 disperses throughout the cell, while Nup96 remains at the nuclear periphery. As cells exit mitosis, Nup49 reassociates with the nuclear periphery of the G1 nuclei (see movie S1 in the supplemental material). Bar, ∼4 μm. (B) Schematic model of an A. nidulans NPC during late G2 and mitosis. In late G2, FG-repeat Nups (red) occupy the central channel of the NPC, restricting passive diffusion and helping to facilitate active transport. Accumulation and activation of the NIMA kinase during G2 trigger the dispersal of central channel Nups, while core Nups (green) remain associated with the nuclear envelope. Putative phosphorylation of Nups by NIMA is indicated. Opening of the NPC central channel compromises active transport and also allows passive diffusion through the NPC. This allows equilibration between the cytoplasm and the nucleus and thus tubulin to gain access to nuclei specifically during mitosis.
Analysis of the predicted location of each of the mitotically dispersed Nups within the NPC structure shed light on what was occurring. All of the FG-repeat Nups predicted to locate to the NPC central channel disperse from the NPC during mitosis, while Nups predicted to be part of the core ring structure and/or to anchor the NPC in the nuclear envelope remain (7, 33). The model predicted by these studies suggests that the central channel of the NPC is open during mitosis in A. nidulans, allowing passive diffusion in and out of nuclei (Fig. 1B). In addition, because the FG-repeat Nups help facilitate active nucleocytoplasmic transport, this model predicts that active transport is not occurring during mitosis in A. nidulans. This model is further supported by the mitosis-specific loss of the cytoplasmic localization of Ran-GAP, which is required to establish a gradient of Ran-GTP across the nuclear envelope that is critical for active transport (7).
These data indicate that A. nidulans undergoes an evolutionary intermediate form of mitosis, which is neither completely closed, because the nuclear envelope is permeable, nor completely open, because the nuclear envelope is intact. The model in which an open NPC central channel makes the nuclear envelope permeable during A. nidulans mitosis helps to explain several previous observations. The early work of Robinow and Caten with A. nidulans, using phase-contrast microscopy, described the loss of demarcation between the cytoplasm and the nuclei as cells entered mitosis, suggesting an equilibration across the nuclear envelope (38). Other work has demonstrated that fluorescent constructs targeted to either the nucleus or the cytoplasm during interphase disperse throughout the cell specifically during mitosis (48; C. P. C. De Souza and S. A. Osmani, unpublished data). Perhaps more functionally significant is the work of Ovechkina and colleagues demonstrating that depolymerized tubulin is excluded from nuclei during interphase but enters nuclei specifically during mitosis in A. nidulans (35).
Regulating nuclear pore complex disassembly.
The genetic interaction between the NIMA kinase and the SONBnNup98 and SONAGle2 Nups strongly suggests that NIMA has a function at the NPC during the G2/M transition (6, 55). This is further supported by the dynamic cell cycle-dependent localization of the NIMA kinase. NIMA accumulates in the cytoplasm in G2 and translocates to nuclei upon mitotic entry, during which time it becomes hyperphosphorylated, further increasing its activity (56). Notably, NIMA is conspicuous at the nuclear periphery during this translocation (7), a feature not shared with other proteins, such as tubulin (35), which enter the nucleus during this window of the cell cycle. Therefore, NIMA is in the right place at the right time to be a Nup kinase, and the localization of dominant negative NIMA constructs strengthens this argument. It has been known for some time that dominant negative NIMA constructs cause a delay in the G2/M transition when expressed either in A. nidulans (25) or in human cells (24). More recent work demonstrates that these dominant negative constructs colocalize with the NPC during this G2 delay, suggesting that they are interfering with endogenous NIMA function at this locale (7).
Biochemical evidence supports the genetic data indicating that NIMA targets the SONBnNup98/SONAGle2 complex during mitotic entry. SONBnNup98 is heavily phosphorylated during mitosis, and both its phosphorylation and its dispersal from the NPC are dependent upon NIMA activation (7). Furthermore, ectopic NIMA expression is sufficient to trigger inappropriate dispersal of SONBnNup98 from the NPC during S-phase arrest (7). Such NIMA expression also dramatically modifies the transport properties of the NPC as nuclear-targeted constructs are released throughout the cell and depolymerized tubulin can enter nuclei (7). Therefore, NIMA expression is sufficient to convert interphase NPCs to a partially disassembled, open mitotic-like state.
Collectively, these studies provide strong evidence that NPC disassembly at the G2/M transition is regulated by Nup phosphorylation carried out by the NIMA kinase (Fig. 1B). However, mitotic entry in A. nidulans requires activation of both the NIMA and the Cdk1/cyclin B kinases (34), and inactivation of either kinase prevents mitotic NPC disassembly, suggesting that they cooperate to regulate disassembly of the NPC (7). The relative contributions of NIMA, Cdk1/cyclin B, and other mitotic kinases in regulating NPC disassembly remain an area for further studies.
Do other organisms undergo partial NPC disassembly?
At present, it is not known how many other members of the eukaryotic kingdom maintain a partially disassembled NPC during mitosis. However, similarly to A. nidulans (38), an apparent equilibration between the cytoplasm and nuclei during closed mitosis also occurs in other fungi, including Ceratocystis fagacearum, Fusarium oxysporum (1), Fusarium verticillioides, and Magnaporthe grisea (3). This suggests that increasing the permeability of the nuclear envelope during mitosis by partial NPC disassembly may occur in many fungi undergoing what were previously thought to be completely closed mitoses.
Interestingly, NPC disassembly occurs in a stepwise manner in higher eukaryotes, and it is noteworthy that the FG-repeat Nups disassemble prior to the core Nups (15). This means there is a period of time in which FG-repeat Nups have dispersed from the central channel of the NPC but in which the nuclear envelope is still intact. This suggests that an open conduit through the NPC may exist during the initial stages of entry into open mitoses. Indeed, in starfish oocytes, nuclear envelope permeability increases in early prophase when FG-repeat Nups disperse from NPCs which still contain other Nups (22). This may be significant in terms of cell cycle regulation, given that nuclear accumulation of the mitotic regulator Cdk1/cyclin B occurs prior to nuclear envelope breakdown in many cell types, including starfish oocytes and mammalian cells (12, 22, 37).
The onset of NPC disassembly is one of the earliest aspects of mitosis, occurring before detectable DNA condensation or spindle formation in A. nidulans and humans. However, little is known about the mechanisms which regulate this process, although the mitosis-specific phosphorylation of many Nups suggests that kinases are involved (14, 26, 29). As mentioned above, the NIMA kinase is a strong candidate for the regulation of this event in A. nidulans. Identifying the targets within the NPC that are potentially phosphorylated by NIMA will provide insights into how the NPC is mitotically disassembled. Interestingly, the conserved p62 Nup (Nsp1) disperses from nuclei when NIMA is expressed in vertebrate cells (24). It remains to be seen if any of the 11 NIMA-related kinases in the human genome (31) regulate NPC function during mitotic entry, but this might be a profitable avenue for future research.
MITOTIC ENTRY THROUGH LOCALIZED NUCLEAR ENVELOPE BREAKDOWN
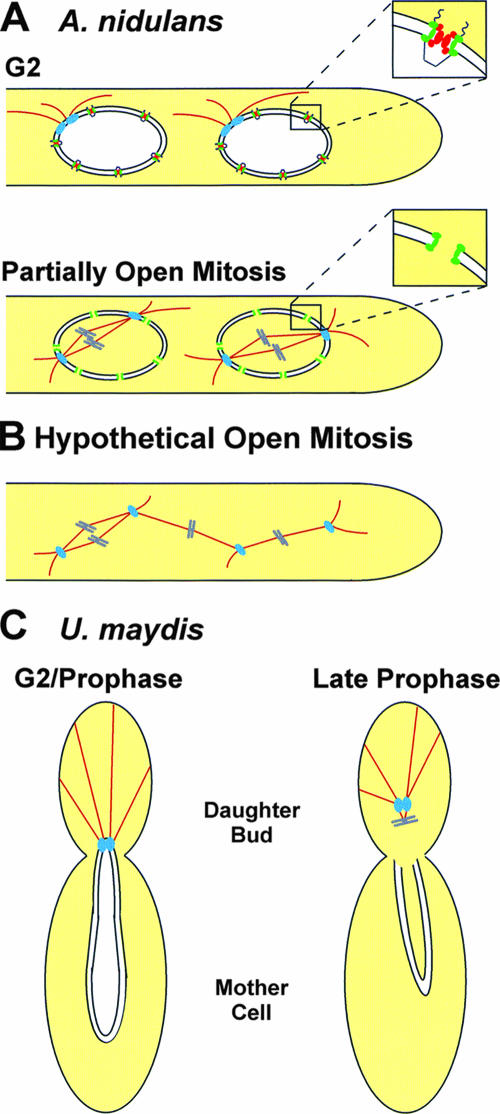
The most dramatic way to open mitosis is to break down the nuclear envelope, as occurs during open mitosis. However, this is not always an all-or-nothing process. For example, in Caenorhabditis elegans syncytial embryos, localized breakdown of the nuclear envelope occurs near the centrosomes and likely allows spindle formation, even though complete NPC disassembly and nuclear envelope breakdown do not occur until anaphase (21). Another striking mechanism for achieving mitotic entry occurs during the yeast-like phase of the dimorphic plant pathogen Ustilago maydis (32, 47). These uninucleate cells undergo budding to generate two cells, each with one daughter nucleus. However, unlike the budding yeast S. cerevisiae, which segregates DNA on a spindle formed in the mother cell, U. maydis chromosomes migrate into the daughter bud where the spindle is subsequently formed (Fig. 2C) (47). Moreover, the nuclear envelope is stripped from DNA as the mitotic spindle forms in the daughter bud, and therefore U. maydis yeast-like cells undergo a form of open mitosis (32, 47). Steinberg and colleagues have begun to identify the mechanisms regulating this process by following the localization of endogenously tagged proteins and manipulating mitotic progression. Nuclear migration is mediated by microtubules nucleated from spindle pole bodies at one end of the nucleus. Together with the dynein molecular motor, these microtubules pull the tip of the nucleus into the daughter bud during late G2 and prophase (Fig. 2C) (46). As the spindle forms, the nuclear envelope breaks down locally in the vicinity of the spindle pole bodies, leaving most of the nuclear envelope in the mother cell (Fig. 2C) (47). Straube et al. have shown that dynein-mediated nuclear migration is required to strip the nuclear envelope from chromosomal DNA, helping to generate the open mitosis in U. maydis (47). This process has similarities to mammalian mitosis, in which dynein facilitates microtubule-based tearing of the nuclear envelope during mitotic entry (40).
FIG. 2.
Variant forms of mitosis change nuclear envelope permeability by different mechanisms. (A) Two A. nidulans nuclei in a common cytoplasm. During G2, the spindle pole bodies (blue) nucleate microtubules in the cytoplasm. The NPC is composed of core Nups (green) and FG-repeat Nups (red), which occupy the NPC central channel and restrict diffusion. Upon mitotic entry, FG-repeat Nups disperse from the central channel, allowing nuclear entry of tubulin and thus spindle formation. Insets show a magnification of a single NPC. Note that the nuclear envelope may prevent inappropriate microtubule connections between the spindle pole bodies and the chromosomes of the two nuclei which are in mitosis at the same time. (B) An illustration of the predicted outcome of an A. nidulans cell undergoing a hypothetical open mitosis. As mitosis is no longer enclosed by the nuclear envelope, microtubules can potentially connect to chromosomes from either nucleus. (C) Mitotic entry during the yeast-like budding phase of U. maydis. During late G2 and early prophase, microtubules pull the tip of the nucleus into the daughter bud in a dynein-dependent manner. Upon mitotic entry, the nuclear envelope breaks down near the spindle pole bodies, and the spindle begins to form in the daughter bud, while most of the nuclear envelope remains in the mother cell. This localized breakdown of the nuclear envelope may help facilitate nuclear envelope permeability. Based on the model depicted by Straube et al. (adapted from reference 47 with permission of the publisher).
Remarkably, experimental evidence indicates that nuclear envelope removal may not be required for successful mitosis in U. maydis and suggests that this organism can also undergo closed mitosis. For instance, when cell wall synthesis is inhibited, bud formation is prevented, and the nucleus enters mitosis within the mother cell (47). Surprisingly, these nuclei undergo a form of closed mitosis as the spindle forms within a nuclear envelope which often appears intact. Similar closed mitoses can also be induced in budded cells if migration of the nucleus into the daughter bud is prevented by drug treatment to depolymerize microtubules or if dynein function is compromised (47). While these experiments helped the authors determine that nuclear migration is involved in nuclear envelope removal during U. maydis open mitosis, they also indicate that this organism can enter an apparently closed mitosis. At present, these experiments have not addressed whether U. maydis cells entering such closed mitoses generate viable daughter cells. However, Straube et al. were also able to prevent nuclear envelope removal by deleting the mitotic exit network component tem1 (ras3 in U. maydis) gene (47). In this case, nuclear migration was normal, and mitosis occurred in the daughter bud. Notably, however, 21% of Δras3 cells with mitotic spindles had a nuclear envelope which appeared intact. As the ras3 deletion mutant is viable, this argues that U. maydis can enter a successful closed mitosis. Interestingly, deletion of other nonessential regulators of mitotic exit in U. maydis similarly results in cells entering an apparently closed mitosis (41). Therefore, while the default mechanism allowing mitotic entry during the yeast-like phase of U. maydis is localized nuclear envelope breakdown, this organism may also be able to complete a closed mitosis.
The ability of U. maydis to enter a closed mitosis suggests that mitosis-specific modifications of NPC function can occur during mitotic entry in this organism to allow spindle formation within the nucleus. NPC function is also likely modified during the initial stages of U. maydis open mitosis as nuclear-targeted constructs begin to disperse from the nucleus while the nuclear envelope is intact. However, it remains to be seen if the U. maydis NPC undergoes mitosis-specific transport modifications similar to those of S. cerevisiae or partially disassembles, similarly to that of A. nidulans.
WHY HAVE VARIATIONS OF MITOSIS EVOLVED?
Why have organisms evolved such seemingly different mechanisms to accomplish mitosis? Evolution dictates that mitosis should occur in its most efficient form. However, different organisms and cell types have different obstacles to overcome to accomplish successful mitosis. To gain better insight into variant mitoses, it is necessary to consider the biology of these different cells.
Location of the microtubule organizing center.
The location of the microtubule organizing center (MTOC) in different cell types provides some clues as to whether the nuclear envelope needs to be broken down during mitosis. The mitotic MTOC of fungi is the spindle pole body, which is embedded in the nuclear envelopes of A. nidulans, S. cerevisiae, and many other fungi. As spindle pole bodies can nucleate microtubules from either their cytoplasmic or nuclear face, it is not essential to break down the nuclear envelope in order to form a mitotic spindle. Rather, spindle pole bodies need only to change their site of microtubule nucleation from the cytoplasmic face during interphase to the nuclear face during mitosis. One way to regulate this is to restrict when tubulin can enter nuclei during the cell cycle. For example, in A. nidulans, nuclear access of depolymerized tubulin is restricted to mitosis (35), when nuclear envelope permeability increases due to partial NPC disassembly (7). This would fulfill the need of A. nidulans to rapidly form and elongate its spindle during its short period of mitosis (∼5 min). In contrast, a rapid nuclear influx of tubulin does not occur during the S. cerevisiae cell cycle, in which the spindle begins to form in S phase and is present throughout most of the cell cycle. Rather, nuclear levels of tubulin increase gradually as the spindle elongates during cell cycle progression. Therefore, a mechanism which would fine-tune the nuclear uptake of tubulin is required in S. cerevisiae, rather than a short period of rapid uptake. This may explain why S. cerevisiae does not disassemble its NPCs but presumably fine-tunes nuclear uptake of tubulin by modifying nuclear transport. Therefore, different requirements for the nuclear uptake of tubulin based upon the time the mitotic spindle is present during the cell cycle may have resulted in the differences in mitotic NPC regulation between A. nidulans and S. cerevisiae. It will be interesting to see if these concepts can be extended to other fungi which maintain their MTOCs in intact nuclear envelopes during mitosis but which have significantly different rates of mitotic spindle formation. In this regard, it is worth noting that Candida albicans behaves similarly to S. cerevisiae in that it maintains a nuclear spindle for most of its cell cycle and maintains an intact NPC during mitosis (10).
In many cells undergoing an open mitosis, the centrosome acts as the MTOC during interphase and mitosis. Given that the centrosome is cytoplasmic while the DNA is nuclear, it is necessary to break down the nuclear envelope in order to form a spindle which can interact with chromosomes. Notably, however, complete breakdown of the nuclear envelope is not necessary for spindle formation, as illustrated by the mitoses of C. elegans (21) and Drosophila melanogaster (36) syncytial embryos. In these organisms, partial breakdown of the nuclear envelope in the vicinity of the centrosomes is sufficient to allow microtubules to capture chromosomes. Nonetheless, the presence of a cytoplasmic MTOC during interphase can generally predict that an organism will undergo some form of nuclear envelope breakdown during mitotic entry. One notable exception to this rule is the fission yeast Schizosaccharomyces pombe. Intriguingly, the S. pombe spindle pole body is cytoplasmic during interphase but embeds in the nuclear envelope during mitotic entry when the spindle forms within an intact nuclear envelope (8, 54). Therefore, relocation of the MTOC to the cytoplasm during interphase does not always predict that a cell will undergo complete nuclear envelope breakdown.
Nuclei in syncytia.
Many organisms contain multiple nuclei within a common cytoplasm, adding further obstacles to achieving successful mitoses. These nuclei are often present at a high density, yet it is vital to ensure that the spindle apparatus attaches correctly to the kinetochores of chromosomes from the appropriate nucleus. One way to achieve this is for nuclei within a common cytoplasm to undergo asynchronous mitoses. In this scenario, because only one nucleus is in mitosis at any given time, microtubules can attach only to chromosomes from the correct nucleus. However, many organisms with syncytial nuclei undergo synchronous or parasynchronous mitoses, resulting in multiple nuclei undergoing mitosis at the same time in the same cytoplasm (13). Another way to prevent inappropriate attachment of microtubules to chromosomes of the incorrect nucleus is to enclose each spindle within its own nuclear envelope. For example, mitosis in A. nidulans occurs in a parasynchronous wave along hyphae, resulting in several nuclei being in mitosis at the same time (Fig. 2A). However, the presence of a nuclear envelope during mitotic entry restricts the attachment of spindle microtubules to only chromosomes within that nucleus (Fig. 2A). If there was no nuclear envelope under these circumstances, microtubules could potentially attach to chromosomes from the incorrect nucleus, as illustrated in Fig. 2B. Such inappropriate microtubule attachments would result in massive missegregation of chromosomes, not fulfilling the mitotic mission, and catastrophic consequences for the organism's survival.
During the early development of complex multicellular organisms such as Drosophila, many nuclei undergo synchronous mitoses within a syncytial cytoplasm. The consequence of chromosomal missegregation in these cells would be particularly disastrous, given the different developmental fates of these nuclei. It is therefore noteworthy that in these cells, complete nuclear envelope breakdown does not occur until after metaphase, when spindle microtubules have attached to the appropriate kinetochores (36). Therefore, evolution may have dictated that nuclei in syncytia either do not completely break down their nuclear envelope until a metaphase spindle has formed or maintain their nuclear envelope throughout mitosis.
Is switching mitotic modes the best of both worlds?
The slime mold Physarum polycephalum can exist as a uninucleate amoeba but also forms a syncytial plasmodium containing many nuclei. During the uninucleate phase of its life cycle, mitosis is open, but remarkably, this organism switches to closed mitosis during the plasmodial phase of its life cycle (44, 49). As described above, undergoing closed mitosis in such syncytia restricts microtubules to attaching to only the appropriate chromosomes by maintaining the spindle apparatus and chromosomes within a nuclear envelope. This may be particularly important given the highly synchronous nature of mitosis within the P. polycephalum syncytium (9, 49). This precedent suggests that other organisms may utilize different forms of mitosis at different stages of their life cycle or in different cell types, depending on which form of mitosis is the most advantageous. Perhaps the ability of U. maydis to enter mitosis with or without an intact nuclear envelope (46) reflects its dimorphic nature. During pathogenic development, U. maydis forms filamentous dikaryons, each containing two genetically distinct nuclei in a common cytoplasm. As with other basidiomycetes (19, 50), the nuclei in these dikaryons undergo synchronous mitoses in close proximity (42). Therefore, we suggest that U. maydis may undergo closed mitosis during dikaryotic filamentous growth to ensure that spindle pole body-nucleated microtubules attach to the kinetochores of the appropriate nucleus. This may explain why during the yeast-like phase of its life cycle U. maydis can undergo closed mitosis even though mitosis is usually open in these cells (46). It will be interesting to see how many other organisms undergo different forms of mitosis in different cell types, but we predict this phenomenon may be more widespread than currently realized.
CONCLUSIONS AND PERSPECTIVES
Mitosis is fundamental to all eukaryotic life, yet many variations of the way in which mitosis is accomplished have evolved in nature. However, all of these mitotic variations allow the organism to achieve the goal of a successful mitosis, the faithful segregation of chromosomes. In this minireview, we have considered why different organisms and cell types may have evolved variant mitoses.
Many fungi achieve successful synchronous mitoses of multiple nuclei in a common cytoplasm by confining each spindle within a complete nuclear envelope. In these closed mitoses, nuclear microtubules emanate from spindle pole bodies embedded within the nuclear envelope. To form a spindle, these organisms must relocate tubulin from the cytoplasm to the nucleus and utilize the NPC to do so. Nuclear access of tubulin through the NPC can occur either by increasing the permeability of the NPC or by modifying active transport through the NPC. We propose that organisms which rapidly form a spindle inside the nuclear envelope utilize partial NPC disassembly to achieve a rapid nuclear influx of tubulin. Other organisms that maintain a nuclear spindle for most of the cell cycle do not disassemble their NPCs but undergo completely closed mitosis. These organisms likely modify nuclear transport to allow tubulin into the nucleus. This may better facilitate the relatively slow spindle elongation which occurs in these organisms by fine-tuning nuclear levels of tubulin.
Clearly, organisms with cytoplasmic centrosomes need to break down their nuclear envelope to allow microtubules access to chromosomes and, therefore, undergo open mitosis. However, if nuclear envelope breakdown occurs in a cytoplasm containing multiple nuclei synchronously undergoing mitosis, centrosomally nucleated microtubules could potentially interact with chromosomes from several different nuclei. To help prevent this, such organisms initially restrict nuclear envelope breakdown to the areas adjacent to centrosomes. The remaining nuclear envelope helps to shelter chromosomes, preventing them from attaching to microtubules nucleated from the inappropriate centrosomes. Therefore, we propose that in syncytia, delay of complete nuclear envelope breakdown until after metaphase spindle formation provides a mechanism with which to prevent microtubules from interacting with chromosomes of inappropriate nuclei.
While considering that the biology of different cells can begin to explain “why” variant mitoses may have evolved, a far greater challenge lies ahead in determining mechanistically “how” variant mitoses are regulated. This will be particularly interesting in the case of organisms which undergo different forms of mitosis in different cell types. In these organisms, the same cell cycle regulators must be able to orchestrate the different forms of mitosis. No doubt the biology of mitosis still has many mysteries to reveal and there are many lessons still to be learned.
Acknowledgments
We thank all members of the Osmani laboratory and Berl Oakley (Ohio State University) for helpful discussions on the topics discussed in this review. We thank Helmut Sauer (Texas A&M University) for interesting discussions regarding P. polycephalum mitosis.
We acknowledge support from National Institutes of Health grant GM 042564 to S.A.O. and a National Research Service Award T32 fellowship to C.P.C.D.
Footnotes
Published ahead of print on 27 July 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aist, J. R. 1969. The mitotic apparatus in fungi, Ceratocystis fagacearum and Fusarium oxysporum. J. Cell Biol. 40:120-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aist, J. R., and P. H. Williams. 1972. Ultrastructure and time course of mitosis in the fungus Fusarium oxysporum. J. Cell Biol. 55:368-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourett, T. M., J. A. Sweigard, K. J. Czymmek, A. Carroll, and R. J. Howard. 2002. Reef coral fluorescent proteins for visualizing fungal pathogens. Fungal Genet. Biol. 37:211-220. [DOI] [PubMed] [Google Scholar]
- 4.Copeland, C. S., and M. Snyder. 1993. Nuclear pore complex antigens delineate nuclear envelope dynamics in vegetative and conjugating Saccharomyces cerevisiae. Yeast 9:235-249. [DOI] [PubMed] [Google Scholar]
- 5.Dasso, M. 2002. The Ran GTPase: theme and variations. Curr. Biol. 12:R502-R508. [DOI] [PubMed] [Google Scholar]
- 6.De Souza, C. P., K. P. Horn, K. Masker, and S. A. Osmani. 2003. The SONBNUP98 nucleoporin interacts with the NIMA kinase in Aspergillus nidulans. Genetics 165:1071-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Souza, C. P., A. H. Osmani, S. B. Hashmi, and S. A. Osmani. 2004. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14:1973-1984. [DOI] [PubMed] [Google Scholar]
- 8.Ding, R., K. L. McDonald, and J. R. McIntosh. 1993. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 120:141-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ducommun, B., Y. Tollon, M. Gares, D. Beach, and M. Wright. 1990. Cell cycle regulation of p34cdc2 kinase activity in Physarum polycephalum. J. Cell Sci. 96:683-689. [DOI] [PubMed] [Google Scholar]
- 10.Finley, K. R., and J. Berman. 2005. Microtubules in Candida albicans hyphae drive nuclear dynamics and connect cell cycle progression to morphogenesis. Eukaryot. Cell 4:1697-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frey, S., R. P. Richter, and D. Gorlich. 2006. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314:815-817. [DOI] [PubMed] [Google Scholar]
- 12.Gabrielli, B. G., C. P. De Souza, I. D. Tonks, J. M. Clark, N. K. Hayward, and K. A. Ellem. 1996. Cytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cells. J. Cell Sci. 109:1081-1093. [DOI] [PubMed] [Google Scholar]
- 13.Gladfelter, A. S. 2006. Nuclear anarchy: asynchronous mitosis in multinucleated fungal hyphae. Curr. Opin. Microbiol. 9:547-552. [DOI] [PubMed] [Google Scholar]
- 14.Glavy, J. S., A. N. Krutchinsky, I. M. Cristea, I. C. Berke, T. Boehmer, G. Blobel, and B. T. Chait. 2007. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107-160 subcomplex. Proc. Natl. Acad. Sci. USA 104:3811-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hase, M. E., and V. C. Cordes. 2003. Direct interaction with nup153 mediates binding of tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell 14:1923-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heath, I. B. 1980. Variant mitoses in lower eukaryotes: indicators of the evolution of mitosis? Int. Rev. Cytol. 64:1-80. [DOI] [PubMed] [Google Scholar]
- 17.Hetzer, M. W., T. C. Walther, and I. W. Mattaj. 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21:347-380. [DOI] [PubMed] [Google Scholar]
- 18.Iouk, T., O. Kerscher, R. J. Scott, M. A. Basrai, and R. W. Wozniak. 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159:807-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasa, M., S. Tanabe, and T. Kamada. 1998. The two nuclei in the dikaryon of the homobasidiomycete Coprinus cinereus change position after each conjugate division. Fungal Genet. Biol. 23:110-116. [DOI] [PubMed] [Google Scholar]
- 20.Kerscher, O., P. Hieter, M. Winey, and M. A. Basrai. 2001. Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation. Genetics 157:1543-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, K. K., Y. Gruenbaum, P. Spann, J. Liu, and K. L. Wilson. 2000. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol. Biol. Cell 11:3089-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenart, P., G. Rabut, N. Daigle, A. R. Hand, M. Terasaki, and J. Ellenberg. 2003. Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J. Cell Biol. 160:1055-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim, R. Y., N. P. Huang, J. Koser, J. Deng, K. H. Lau, K. Schwarz-Herion, B. Fahrenkrog, and U. Aebi. 2006. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 103:9512-9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, K. P., and T. Hunter. 1995. Evidence for a NIMA-like mitotic pathway in vertebrate cells. Cell 81:413-424. [DOI] [PubMed] [Google Scholar]
- 25.Lu, K. P., and A. R. Means. 1994. Expression of the non-catalytic domain of the NIMA kinase causes a G2 arrest in Aspergillus nidulans. EMBO J. 13:2103-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macaulay, C., E. Meier, and D. J. Forbes. 1995. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J. Biol. Chem. 270:254-262. [DOI] [PubMed] [Google Scholar]
- 27.Makhnevych, T., C. P. Lusk, A. M. Anderson, J. D. Aitchison, and R. W. Wozniak. 2003. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell 115:813-823. [DOI] [PubMed] [Google Scholar]
- 28.Marelli, M., J. D. Aitchison, and R. W. Wozniak. 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143:1813-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, M. W., M. R. Caracciolo, W. K. Berlin, and J. A. Hanover. 1999. Phosphorylation and glycosylation of nucleoporins. Arch. Biochem. Biophys. 367:51-60. [DOI] [PubMed] [Google Scholar]
- 30.Morris, N. R. 1976. Mitotic mutants of Aspergillus nidulans. Genet. Res. 26:237-254. [DOI] [PubMed] [Google Scholar]
- 31.O'Connell, M. J., M. J. Krien, and T. Hunter. 2003. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 13:221-228. [DOI] [PubMed] [Google Scholar]
- 32.O'Donnell, K. L., and J. T. McLaughlin. 1984. Postmeiotic mitosis, basidiospore development, and septation in Ustilago maydis. Mycologia 76:486-502. [Google Scholar]
- 33.Osmani, A. H., J. Davies, H. L. Liu, A. Nile, and S. A. Osmani. 2006. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell 17:4946-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osmani, A. H., S. L. McGuire, and S. A. Osmani. 1991. Parallel activation of the NIMA and p34cdc2 cell cycle-regulated protein kinases is required to initiate mitosis in A. nidulans. Cell 67:283-291. [DOI] [PubMed] [Google Scholar]
- 35.Ovechkina, Y., P. Maddox, C. E. Oakley, X. Xiang, S. A. Osmani, E. D. Salmon, and B. R. Oakley. 2003. Spindle formation in Aspergillus is coupled to tubulin movement into the nucleus. Mol. Biol. Cell 14:2192-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paddy, M. R., H. Saumweber, D. A. Agard, and J. W. Sedat. 1996. Time-resolved, in vivo studies of mitotic spindle formation and nuclear lamina breakdown in Drosophila early embryos. J. Cell Sci. 109:591-607. [DOI] [PubMed] [Google Scholar]
- 37.Pines, J., and T. Hunter. 1991. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol. 115:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinow, C. F., and C. E. Caten. 1969. Mitosis in Aspergillus nidulans. J. Cell Sci. 5:403-431. [DOI] [PubMed] [Google Scholar]
- 39.Rout, M. P., and J. D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593-16596. [DOI] [PubMed] [Google Scholar]
- 40.Salina, D., K. Bodoor, D. M. Eckley, T. A. Schroer, J. B. Rattner, and B. Burke. 2002. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108:97-107. [DOI] [PubMed] [Google Scholar]
- 41.Sandrock, B., C. Bohmer, and M. Bolker. 2006. Dual function of the germinal centre kinase Don3 during mitosis and cytokinesis in Ustilago maydis. Mol. Microbiol. 62:655-666. [DOI] [PubMed] [Google Scholar]
- 42.Scherer, M., K. Heimel, V. Starke, and J. Kamper. 2006. The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell 18:2388-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 44.Solnica-Krezel, L., T. G. Burland, and W. F. Dove. 1991. Variable pathways for developmental changes of mitosis and cytokinesis in Physarum polycephalum. J. Cell Biol. 113:591-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart, M. 2007. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8:195-208. [DOI] [PubMed] [Google Scholar]
- 46.Straube, A., W. Enard, A. Berner, R. Wedlich-Soldner, R. Kahmann, and G. Steinberg. 2001. A split motor domain in a cytoplasmic dynein. EMBO J. 20:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Straube, A., I. Weber, and G. Steinberg. 2005. A novel mechanism of nuclear envelope break-down in a fungus: nuclear migration strips off the envelope. EMBO J. 24:1674-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suelmann, R., N. Sievers, and R. Fischer. 1997. Nuclear traffic in fungal hyphae: in vivo study of nuclear migration and positioning in Aspergillus nidulans. Mol. Microbiol. 25:757-769. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka, K. 1973. Intranuclear microtubule organizing center in early prophase nuclei of the plasmodium of the slime mold, Physarum polycephalum. J. Cell Biol. 57:220-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torralba, S., A. G. Pisabarro, and L. Ramirez. 2004. Immunofluorescence microscopy of the microtubule cytoskeleton during conjugate division in the dikaryon Pleurotus ostreatus N001. Mycologia 96:41-51. [PubMed] [Google Scholar]
- 51.Tran, E. J., and S. R. Wente. 2006. Dynamic nuclear pore complexes: life on the edge. Cell 125:1041-1053. [DOI] [PubMed] [Google Scholar]
- 52.Weis, K. 2003. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112:441-451. [DOI] [PubMed] [Google Scholar]
- 53.Wente, S. R. 2000. Gatekeepers of the nucleus. Science 288:1374-1377. [DOI] [PubMed] [Google Scholar]
- 54.West, R. R., E. V. Vaisberg, R. Ding, P. Nurse, and J. R. McIntosh. 1998. cut11(+): a gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell 9:2839-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, L., S. A. Osmani, and P. M. Mirabito. 1998. A role for NIMA in the nuclear localization of cyclin B in Aspergillus nidulans. J. Cell Biol. 141:1575-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye, X. S., G. Xu, P. T. Pu, R. R. Fincher, S. L. McGuire, A. H. Osmani, and S. A. Osmani. 1995. The NIMA protein kinase is hyperphosphorylated and activated downstream of p34cdc2/cyclin B: coordination of two mitosis promoting kinases. EMBO J. 14:986-994. [DOI] [PMC free article] [PubMed] [Google Scholar]