Abstract
Among eukaryotes, only slime molds, fungi, and plants contain signal transduction phosphorelay systems. In filamentous fungi, multiple sensor kinases appear to use a single histidine-containing phosphotransfer (HPt) protein to relay signals to two response regulators (RR). In Aspergillus nidulans, the RR SskA mediates activation of the mitogen-activated protein kinase SakA in response to osmotic and oxidative stress, whereas the functions of the RR SrrA were unknown. We used a genetic approach to characterize the srrA gene as a new member of the skn7/prr1 family and to analyze the roles of SrrA in the phosphorelay system composed of the RR SskA, the HPt protein YpdA, and the sensor kinase NikA. While mutants lacking the HPt protein YpdA are unviable, mutants lacking SskA (ΔsskA), SrrA (ΔsrrA), or both RR (ΔsrrA ΔsskA) are viable and differentially affected in osmotic and oxidative stress responses. Both RR are involved in osmostress resistance, but ΔsskA mutants are more sensitive to this stress, and only SrrA is required for H2O2 resistance and H2O2-mediated induction of catalase CatB. In contrast, both RR are individually required for fungicide sensitivity and calcofluor resistance and for normal sporulation and conidiospore viability. The ΔsrrA and ΔsskA sporulation defects appear to be related to decreased mRNA levels of the key sporulation gene brlA. In contrast, conidiospore viability defects do not correlate with the activity of the spore-specific catalase CatA. Our results support a model in which NikA acts upstream of SrrA and SskA to transmit fungicide signals and to regulate asexual sporulation and conidiospore viability. In contrast, NikA appears dispensable for osmotic and oxidative stress signaling. These results highlight important differences in stress signal transmission among fungi and define a phosphorelay system involved in oxidative and osmotic stress, cell wall maintenance, fungicide sensitivity, asexual reproduction, and spore viability.
The ability to respond to a vast array of environmental signals is vital for the growth and survival of microorganisms. In prokaryotic cells, the sensing and processing of these signals rely largely on two-component signal transduction pathways that depend on histidine and aspartyl residues as phosphoryl group donors and acceptors. The prototypical two-component signal transduction pathway comprises two protein components, a sensor histidine kinase (HK) and a response regulator (RR). A typical HK consists of a signal-sensing (or input) domain and a transmitter domain that has a conserved kinase core with an invariant histidine residue, whereas a typical RR consists of an N-terminal receiver domain that has an invariant aspartate residue and a C-terminal output domain. Signal reception by the HK stimulates ATP-dependent autophosphorylation at the conserved His residue in its transmitter domain. The phosphokinase then donates the phosphoryl group to the conserved Asp residue in the receiver domain of the cognate RR, thereby rendering it functional, generally as a transcriptional regulator. Many two-component systems, however, are more elaborate. For instance, signal transmission may involve hybrid HKs that contain both transmitter and receiver domains, and additional proteins as phosphorelay components (27, 51, 56).
Among eukaryotic cells, phosphorelay systems have been described for slime molds, fungi, and plants (45, 50, 55, 58, 64, 68). Fungal phosphorelay systems consist of one or several hybrid HKs, a histidine-containing phosphotransfer (HPt) protein, and two canonical RRs. For instance, Saccharomyces cerevisiae has a single membrane-associated HK, Sln1; one HPt protein, Ypd1; and two RRs, Ssk1 and Skn7 (also called Pos9). Under normal osmotic conditions, Sln1 is an active kinase that autophosphorylates at the conserved histidine of the transmitter domain. The phosphoryl group is then successively transferred to the conserved aspartate of the Sln1 receiver domain, to the conserved histidine of Ypd1, and finally to the conserved aspartates of Ssk1 and Skn7 (58). Phosphorylation of Ssk1 prevents the activation of the partially redundant mitogen-activated protein kinase (MAPK) kinase kinases Ssk2 and Ssk22. High-osmolarity conditions promote dephosphorylation of phosphorylated Sln1, resulting in accumulation of unphosphorylated Ssk1. Unphosphorylated Ssk1 interacts physically with Ssk2 and Ssk22, leading to their activation and the subsequent phosphorylation of Pbs2 and Hog1 (57). Phosphorylated Hog1, in turn, activates various transcription factors, responsible for the induction of genes required for survival in a hyperosmotic environment (see reference 28 for a review). The other RR, Skn7, is a typical RR with a DNA-binding domain similar to that of the heat shock factor Hsf1 (11) and thus is directly involved in transcriptional regulation. The skn7 gene was identified in different screenings as a high-copy suppressor of mutations affecting cell wall assembly (11) and lethality associated with loss of the transcription factors SBF and MBF (47), as a mutation causing increased sensitivity to oxidative stress (37), and as a high-copy activator of a SLN1-dependent reporter gene (41).
In contrast to S. cerevisiae, the fission yeast Schizosaccharomyces pombe contains not one but three HKs, Mak1, Mak2, and Mak3, in addition to the Ypd1-orthologous HPt protein Mpr1 and the two RRs Mcs4 and Prr1. Although these phosphorelay pathways are similar to the Sln1-Ypd1-Ssk1 system, they are specialized in transmitting oxidative rather than osmotic stress signals. Mak2 and Mak3 (12) have been shown to transmit oxidative stress signals, through Mcs4, to the stress MAPK Spc1/Sty1 (12, 50), which is a Hog1 ortholog. Mak1 appears to regulate Prr1 (12, 54), the Skn7 ortholog. The prr1− mutant is sensitive to H2O2 and cadmium and is defective in the induction of the ctt1+ gene (53), which encodes the only catalase present in this fungus. The analysis of several fungal genomes indicates that the number of HKs in filamentous fungi can range from 11 to 21, and all of them appear to use a single HPt protein to relay signals to two conserved RRs (9, 14). How different environmental signals are perceived and integrated through these proteins is a matter of intense research.
Aspergillus nidulans is closely related to other aspergilli of medical or industrial importance and is a good genetic model with which to study the biology of fungi and other eukaryotes (2, 5, 16, 19, 44, 69, 70, 76). In this fungus, asexual reproduction (conidiation) is induced by environmental signals such as exposure to air (2, 16, 69), nutrient starvation (66), and self-generated signals (39, 43, 65, 67, 70). The mechanisms by which these signals are perceived and transduced are not well understood.
Conidiation implicates the development of specialized structures called conidiophores that are able to produce numerous uninucleated conidiospores. As dormant structures, capable of resisting different types of stress and germinating under favorable conditions, conidiospores are critical for the dispersal of A. nidulans and many other fungi. The formation of conidiophores and conidia is completely dependent on the expression of the brlA gene (1, 3, 16, 66).
The A. nidulans genome (19) and our own analysis predict 15 HKs, 1 HPt protein, and 2 canonical RRs called SskA (17) and SrrA (this work; GenBank accession no. AY168636). Moreover, two additional proteins with putative receiver domains are predicted: AN7572.3, homologous to S. cerevisiae Rim15, which lacks the aspartate that is conserved in canonical RRs, and AN4134.3, which lacks a clear effector domain. The functions of the three HK-encoding genes (tcsA, tcsB, and fphA) have been investigated for this fungus. The tcsA gene encodes a PAS/PAC domain HK that belongs to the class IV HKs (14). Conidiation was reduced in tcsA mutants, a phenotype that was suppressed under high osmolarity or after successive propagation of the strains (73). Mutation of tcsB, the ortholog of yeast sln1, did not produce any clear phenotype (18). Finally, fphA was shown to encode a red-light sensor phytochrome involved in repression of sexual development (8).
A. nidulans SakA, also called HogA (24), is a Hog1 and Spc1/Sty1 ortholog that can replace Spc1 functions in S. pombe and is activated by both oxidative and osmotic stress signals. However, mutants lacking SakA are not sensitive to osmotic stress (34). Recently, it was found that the RR SskA mediates the activation of SakA in response to oxidative and osmotic stress signals (17). We have proposed that oxidative stress plays a central role in determining cell differentiation in fungi and other eukaryotes (5, 25). In this context, we are interested in understanding how filamentous fungi perceive and respond to oxidative and other stress signals. Given the importance of phosphorelay systems in transmitting oxidative stress signals in S. pombe (29) and other fungi (4, 7, 15), we sought to determine the roles of the HK NikA, the HPt protein YpdA, and the RRs SrrA and SskA in A. nidulans stress signal transduction and cell differentiation.
MATERIALS AND METHODS
Cloning of srrA and deletion of ypdA, srrA, sskA, and nikA.
A search of a public A. nidulans cDNA sequence database (59) for genes encoding possible RRs homologous to S. pombe Prr1 and S. cerevisiae Skn7 led to the identification of a 353-bp clone. We used this sequence to design external primers and amplify the corresponding fragment, which was then used to hybridize a chromosome-specific library (10). Cosmids W23C01, W17E02, W17E03, and W4E08, all from chromosome II, hybridized to this probe. Cosmid W23C01 was used as a template to generate the ssrA gene sequence (GenBank accession no. AY168636) by automatic fluorescent dideoxy sequencing in an ABI Prism 310 sequencer from Perkin-Elmer. A srrA replacement construct was generated by double-joint PCR (75) using genomic DNA as a template. First, a 5′ srrA fragment was amplified with primers SrrA5′f and SrrA5′r (see Table 1 for primer sequences). Second, a 3′ srrA fragment was amplified with primers SrrA3′f and SrrA3′r. Third, the A. fumigatus pyrG marker was amplified as a 1,890-bp fragment with primers pyrGreverse and pyrGforward, using plasmid pFNO3 as a template (49). The three PCR fragments were purified, mixed, and subjected to fusion PCR with nested primers SrrAnestf and SrrAnestr. The final 4,949-bp srrA-AfpyrG-srrA cassette was purified with a QIAquick gel purification kit (QIAGEN, Hilden, Germany) and used to transform A. nidulans strains 11035 and CFL3 by protoplast fusion and conidium electroporation (63), respectively.
TABLE 1.
DNA primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| SrrA5′f | TGCCTCTATGCCAGTTCC |
| SrrA5′r | AGGGTGAAGAGCATTGTTTGAGGCCGGAATCAAGGGAAAAAG |
| SrrA3′f | ACGCATCAGTGCCTCCTCGACGCAACGGAAACGATAACC |
| SrrA3′r | GCGCCCCTCCATACAAAA |
| SrrAnestf | AGAGCTTGGGGGAGCAGACG |
| SrrAnestr | GCTCGAATCTGGTCTCAAACG |
| SskA5′f | GCCAGTAACCCTGACACC |
| SskA5′r | ATTTTAATCCCATGTGATCAAACGCAGACGCTTAAGGGGGACAGAA |
| SskA3′f | CGTCACACTCATGTAACGGTTCTGCAGCGAACTGAACTGGAACATTTGG |
| SskA3′r | GGAGCTTAACGCCGATAC |
| SskAnestf | GACAGAATACCCATACAAGGC |
| SskAnestr | CAAAACGCTGATACCCAGTGG |
| Hpt5′f | CTGATTAGGGCTGCTTGG |
| Hpt5′r | GGTGAAGAGCATTGTTTGAGGCTTTGGAGGATTTGGATGGACG |
| Hpt3′f | GCATCAGTGCCTCCTCTCAGACTTATGCGGACTCTCAAGC |
| Hpt3′r | CGATTGTCAGGTGGATGG |
| Hptnestf | TTCGACCCTACCCAGTGC |
| Hptnestr | CGTCAGCGATGATCTCAG |
| PyrGforward | GCCTCAAACAATGCTCTTCACC |
| PyrGreverse | GCTTGAGAGGAGGCATGATGC |
| 5Ribo | CTGGCTCGTTTGATCACATGG |
| 6Ribo | GCGCTGCAGAACCGTTACATG |
| 5DignkuA | GATGCTGTCCTTTTTGCC |
| 3DignkuA | GCTGAAGAGTGCGAGACG |
| 5′ANnik1f | GGGCATGGATGTTGACATCGG |
| 5′ANnik1r | GGTGAAGAGCATTGTTTGAGGCGCTAGAACAGGGGGTGAAGAA |
| 3′ANnik1f | GCATCAGTGCCTCCTCTCAGAAGACCATGATACCATGATACC |
| 3′ANnik1r | GGATTTGTGTGCTTACAGAGT |
| 5′ANnik1nest | GCTTCAGTCCCGTGACATTGC |
| 3′ANnik1nest | GGAACTAAAAGCGTGAGGAGC |
A similar strategy was used to delete the sskA gene by using primers SskA5′f, SskA5′r, SskA3′f, SskA3′r, SskAnestf, and SskAnestr (Table 1). In this case, the A. fumigatus riboB marker was amplified as a 1,943-bp product with primers 5Ribo and 6Ribo by using plasmid pAfriboPstE1Skt(ssp1)-37 as a template. This plasmid contains a PstI-EcoRI fragment containing the Aspergillus fumigatus riboB gene (49) cloned into pBluescript SK(+) and was a generous gift from Berl Oakley's laboratory (Ohio State University). The final 5,101-bp sskA-AfriboB-sskA cassette was purified as described above and used to transform A. nidulans strain 11035 by protoplast fusion. To delete ypdA, we used primers Hpt5′f, Hpt5′r, Hpt3′f, Hpt3′r, Hptnestf, and Hptnestr. The A. fumigatus pyrG marker was amplified as described above, and the final 4,258-bp purified construct, hptA::AfpyrG::hptA, was used to transform strain 11035 by protoplast fusion and conidium electroporation. The nikA gene (AN4479.3) deletion construct nikA::AfpyrG::nikA was generated using primers 5′ANnik1f, 5′ANnik1r, 3′ANnik1f, 3′ANnik1r, 5′ANnik1nest, 3′ANnik1nest, pyrGreverse, and pyrGforward and was used to transform strain 11035.
Strains, media, and growth conditions.
The A. nidulans strains used in this work are listed in Table 2. Genotypes of progeny derived from sexual crosses were initially determined by colony morphology and growth tests on media containing high sorbitol, H2O2, or the fungicide fludioxonil and were confirmed by Southern blot and/or PCR analysis. The presence of the wild-type nkuA+ allele was determined by PCR using primers 5DignkuA and 3DignkuA. All strains were grown in glucose minimal nitrate medium (26) plus supplements. Menadione and fludioxonil were filter sterilized and, like H2O2, added to agar medium at ∼50°C before solidification. H2O2-containing plates were either used the day they were prepared or stored at 4°C for no more than 24 h. Since H2O2 can react with medium components, the actual concentration in plates cannot be estimated. To ensure experimental reproducibility, the same batch of H2O2-containing medium was used for comparing different strains. To test heterokaryon sensitivity under osmotic and oxidative stress, mycelial plugs of the same area (diameter, 0.5 cm) were cut from the borders of growing heterokaryotic colonies and transferred to the testing medium. Induction of catalase CatB by H2O2 and in-gel catalase activity were determined as reported previously (33, 35, 48). Induction of asexual sporulation, sample processing, and brlA Northern blot hybridization were carried out as reported previously (3, 66).
TABLE 2.
Aspergillus nidulans strains used in this study
| Strain | Genotypea | Source |
|---|---|---|
| CLK43 | pabaA1 yA2 veA1 | 34 |
| CLK56 | pabaA1 yA2 catB(−3500)::lacZ veA1 | L. Kawasaki; progeny from TLK42 × CLK43 |
| TOL1 | pabaA1 yA2 ΔargB::trpCΔB ΔsakA::argB trpC801 veA1 | 34 |
| 11035 | pyrG89 pyroA4 ΔnkuA::argB riboB2 veA1* | M. Hynes; 49 |
| CFL3 | pyrG89 pabaA1 yA2 veA1 | F. Lara; progeny from CLK43 × SRF200 |
| SRF200 | pyrG89 ΔargB::trpC pyroA4 veA1 | R. Fischer |
| TΔsrrA-pyrG(ΔnkuA)7 | pyrG89 ΔsrrA::AfpyrG pyroA4 ΔnkuA::argB riboB2 veA1* | This work; 11035 transformed with PCR construct srrA-AfpyrG-srrA |
| TΔsrrA-pyrG9 | pyrG89 pabaA1 yA2 ΔsrrA::AfpyrG veA1 | This work; CFL3 transformed with PCR construct srrA-AfpyrG-srrA |
| TΔsskA-riboB(ΔnkuA)8 | pyrG89 pyroA4 ΔsskA::AfriboB ΔnkuA::argB riboB2 veA1* | This work; 11035 transformed with PCR construct sskA-AfriboB-sskA |
| TΔsrrA-pyrG/ΔsskA-riboB(ΔnkuA)6 | pyrG89 ΔsrrA::AfpyrG pyroA4 ΔsskA::AfriboB ΔnkuA::argB riboB2 veA1* | This work; 11035 transformed with PCR constructs srrA-AfpyrG-srrA and sskA-AfriboB-sskA |
| TΔnikA-pyrG(ΔnkuA)9 | pyrG89 ΔnikA::AfpyrG pyroA4 ΔnkuA::argB riboB2 veA1* | This work; 11035 transformed with PCR construct nikA-AfpyrG-nikA |
| COSΔsrrA03 | pabaA1 yA2 ΔsrrA::AfpyrG veA1 | This work; progeny from TΔsrrA-pyrG9 × TΔsskA-riboB(ΔnkuA)8 |
| COSΔsskA02 | pabaA1 yA2 ΔsskA::AfriboB veA1 | This work; progeny from TΔsskA-riboB(ΔnkuA)8 × CLK56 |
| COSΔsrrA/ΔsskA02 | pabaA1 yA2 ΔsrrA::AfpyrG ΔsskA::AfriboB veA1* | This work; progeny from TΔsrrA-pyrG9 × TΔsskA-riboB(ΔnkuA)8 |
| CIVΔnikA3 | pabaA1 yA2 ΔnikA::AfpyrG veA1* | This work; progeny from TΔnikA-pyrG(ΔnkuA)9 × CLK43 |
*, partial genotype; it may contain argB2.
RESULTS
The srrA gene encodes an RR of the Skn7/Prr1 family.
The S. cerevisiae Skn7 (11, 37, 40) and S. pombe Prr1 (46, 50, 53) RRs are known to control key functions in response to oxidative stress. By searching a public A. nidulans cDNA database (59), we identified a partial cDNA predicting a 100-amino-acid open reading frame (ORF) sharing 66% identity with the conserved DNA-binding domain of S. pombe Prr1. Using a probe based on this sequence, we identified several clones in a chromosome-specific library and selected cosmid W23C01 to generate the genomic sequence previously submitted to GenBank (accession no. AY168636). Since a 556-amino-acid protein deduced from this sequence showed high similarity to Prr1, we assumed a similar function in A. nidulans, and therefore we named the corresponding gene srrA (for stress response regulator A). Although part of the predicted SrrA protein is identical to hypothetical protein AN3688.2, derived from the A. nidulans genome database (http://www.broad.mit.edu/annotation/genome/aspergillus_nidulans/Home.html) (19), AN3688.2 has only 475 amino acids. A more careful analysis of srrA genomic and partial cDNA sequences, as well as protein alignments with orthologs from other fungi, indicates that srrA actually encodes a 627-amino-acid protein, most similar to hypothetical proteins from Aspergillus clavatus and Neosartorya fischeri (see Fig. S1 in the supplemental material). According to this new analysis, our genomic sequence (GenBank accession no. AY168636) is missing 43 bp of coding sequence, corresponding to the last 15 amino acids of SrrA. The intron positions in srrA and the recently corrected A. fumigatus skn7 (Afskn7) (38) gene appear conserved, and the predicted proteins are very similar. However, the Afskn7 corrections are not yet reflected in public databases. The SrrA DNA-binding and receiver domains are well conserved in all fungal orthologs, including S. cerevisiae and S. pombe. However, high overall conservation is observed only for proteins from filamentous fungi. Comparison of the corrected SrrA sequence (see Fig. S1 in the supplemental material) with proteins in GenBank and other public databases suggests that hypothetical orthologs from most filamentous fungi appear incorrectly annotated, since they lack N- and C-terminal regions present in SrrA.
Deletion of genes encoding the HPt protein YpdA and the RR proteins SrrA and SskA.
To analyze the function(s) of the A. nidulans signal transduction pathways that are formed by the phosphotransfer protein YpdA (17) and RRs SrrA and SskA, we attempted to delete the corresponding genes and generate ΔypdA, ΔsrrA, ΔsskA, and ΔsrrA ΔsskA null mutants. The deletion constructs hptA-AfpyrG-hptA (Fig. 1A), srrA-AfpyrG-srrA (see Fig. S2A in the supplemental material), and sskA-AfriboB-sskA (see Fig. S2B in the supplemental material) were generated by double-joint PCR (75) and were used to transform protoplasts or electrocompetent conidia (62, 63) from the A. nidulans nkuA− strain 11035. In this strain, most DNA recombination events occur via homologous recombination (49).
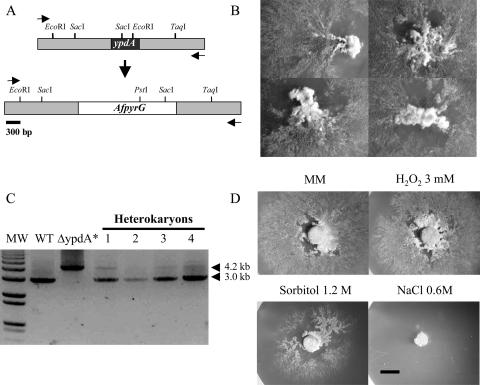
FIG. 1.
Deletion of the ypdA gene in heterokaryons. (A) A ypdA deletion construct based on the AfpyrG gene, used as a selective marker, was generated by double-joint PCR and used to transform strain 11035. The expected integration event results in the replacement of the wild-type ypdA locus (top) by the ΔypdA allele (bottom). (B) PyrG+ heterokaryotic transformants obtained with the ypdA deletion construct. Spores from these heterokaryons were unable to grow in selective medium (data not shown). (C) The presence of the deleted ypdA allele was confirmed by PCR using genomic DNA from the PyrG+ heterokaryons and primers Hptnestf and Hptnestr (arrowheads in panel A). With these primers, wild-type (WT) ypdA generates a product of 3,014 bp using DNA from either WT or heterokaryotic (lanes 1 to 4) strains. The ypdA deletion allele generates a 4,258-bp PCR product, obtained only by using the ypdA deletion construct (ΔypdA*) and heterokaryotic DNA (lanes 1 to 4). Consistent with this, primers Hpt5′f (located outside of the 5′ end of the deletion construct) and PyrGreverse (located in the 3′ end of AfpyrG) generated the expected PCR product only when heterokaryotic DNA was used as a template (data not shown). (D) Mycelial plugs from growing heterokaryon 4 were transferred to a medium containing either 0.6 M NaCl, 1.2 M sorbitol, or 3 mM H2O2 and were incubated for 4 days at 37°C. Bar, 0.5 cm.
Furukawa et al. (17) were unable to obtain ΔypdA mutants by transforming a regular strain using protoplast fusion, a procedure involving the use of a high-osmolarity medium. Here we used an alternative strategy, aimed at maximizing gene-targeting efficiency (nkuA− strain 11035) and avoiding possible high-osmolarity counterselection of the desired mutants (transformation by conidium electroporation). This procedure generated eight PyrG+ transformants, all showing irregular growth and conidiation, characteristic of A. nidulans heterokaryons (Fig. 1B). Conidiospores isolated from these colonies failed to grow in selective medium, suggesting that deletion of ypdA is a lethal event. To confirm this, we extracted DNA from several heterokaryons and used it for PCR analysis. The results in Fig. 1C show that in contrast to wild-type DNA, heterokaryotic DNA generated products of 3 and 4.2 kb, consistent with amplification of both wild-type and ΔypdA alleles. The smaller amount of the ΔypdA PCR product suggests that heterokaryons contain a higher proportion of ypdA+ nuclei. Additional PCR analysis (data not shown) using a ypdA primer external to the deletion construct (Fig. 1A) and primer PyrGreverse generated the expected PCR product only when heterokaryotic DNA was used as a template, further confirming ypdA deletion in the heterokaryons. We conclude that the lack of the HPt protein YpdA very likely results in a lethal event in A. nidulans.
Nevertheless, we explored the sensitivities of ypdA−/ypdA+ heterokaryons to osmotic and oxidative stress conditions by transferring mycelial plugs from growing heterokaryons 1 and 4 to a medium containing either 1 to 3 mM H2O2 or a high sorbitol or NaCl concentration. The responses of both heterokaryons were very similar under all conditions tested. While similar growth was observed in minimal medium (MM) and in the presence of H2O2, growth was decreased in 1.2 M sorbitol and was totally inhibited in 0.6 M NaCl (Fig. 1D). These results indicate that the partial lethality of the ypdA deletion in heterokaryons is increased by high osmolarity but is not affected by high H2O2 levels. To determine the terminal phenotype of the ypdA mutant, spores collected from ypdA−/ypdA+ heterokaryons were germinated in selective liquid medium for 16 h and examined under the microscope. Although all spores failed to germinate under these conditions, two different morphologies were distinguished. About 80% of the conidia were swollen and bigger than the remaining spores (data not shown). In line with the low ratio of the ΔypdA/ypdA+ PCR product in Fig. 1C, this result suggests that the spores that did not swell include ΔypdA (PyrG+) spores and thus the possible ΔypdA terminal phenotype. In any case, it is clear that ΔypdA mutants can be maintained only as heterokaryons and that ΔypdA conidiospores appear unable to form a germ tube.
To delete most of the srrA ORF, including the DNA-binding and receiver domain regions (see Fig. S1 in the supplemental material), we used the srrA-AfpyrG-srrA replacement construct. Several PyrG+ transformants were analyzed by Southern blotting (data not shown), and transformants with the correct gene replacement event [TΔsrrA-pyrG(ΔnkuA)7 and -8 and TΔsrrA-pyrG9] were selected for further experiments. ΔsrrA mutants obtained from sexual crosses were further analyzed by Southern blotting (see Fig. S2A in the supplemental material).
The sskA-AfriboB-sskA construct, designed to delete the entire sskA ORF, generated 11 RiboB+ transformants, 9 of which were screened by Southern blotting. Transformants TΔsskA-riboB(ΔnkuA)7 and -8 were shown to contain the desired ΔsskA gene replacement event (see Fig. S2B in the supplemental material) and were used in further experiments. When both constructs (srrA-AfpyrG-srrA and sskA-AfriboB-sskA) were used to transform strain 11035, eight PyrG+ riboB+ transformants were obtained. Strains TΔsrrA-pyrG/ΔsskA-riboB1(ΔnkuA)6 and -7 were shown to contain the deletions of the srrA and sskA genes and were selected for additional experiments (see Fig. S3A in the supplemental material).
The deletions of srrA and sskA resulted in phenotypes that were clearly distinguishable from the wild-type phenotype (Fig. 2 to 4; see also Fig. S4 in the supplemental material). ΔsrrA mutants formed colonies with irregular borders and with apparent reductions in conidiophore density and conidiospore pigmentation. In addition, the mycelia from ΔsrrA mutants accumulated a pink pigment. ΔsskA mutants showed similar but less accentuated phenotypes, and their colony borders were more regular. ΔsrrA ΔsskA double mutants had a phenotype similar to that displayed by ΔsskA mutants. Sexual crosses between selected ΔsrrA and ΔsskA mutants and wild-type strains showed that the mutant phenotypes segregated in a Mendelian fashion and allowed us to obtain a set of isogenic ΔsrrA, ΔsskA, and ΔsrrA ΔsskA mutants in an nkuA+ background (Table 2). Similar results were obtained using nkuA− mutants derived from strain 11035 and the nkuA+ strains derived from sexual crosses. We verified the lack of srrA and sskA transcripts in ΔsrrA and ΔsskA mutants by Northern blot analysis using strains COSΔsrrA03 and COSΔsskA02 (Table 2), respectively. The srrA and sskA transcripts are present at relatively low levels during growth and asexual development, and it does not appear that the mutation of one gene affects the mRNA accumulation of the other (data not shown).
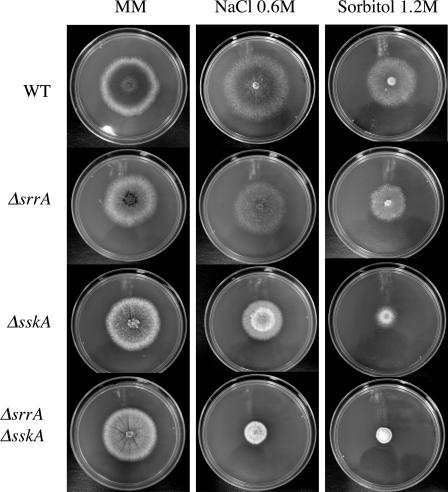
FIG. 2.
ΔsskA and ΔsrrA ΔsskA mutants are sensitive to high-osmolarity stress. Strains 11035 (wild type [WT]), TΔsrrA-pyrG7 (ΔsrrA), TΔsskA-riboB8 (ΔsskA), and TΔsrrA-pyrG/ΔsskA-riboB8 (ΔsrrA ΔsskA) were point inoculated onto supplemented MM plates containing either 0.6 M NaCl or 1.2 M sorbitol and were incubated at 37°C for 5 days. See Table 2 for full genotypes.
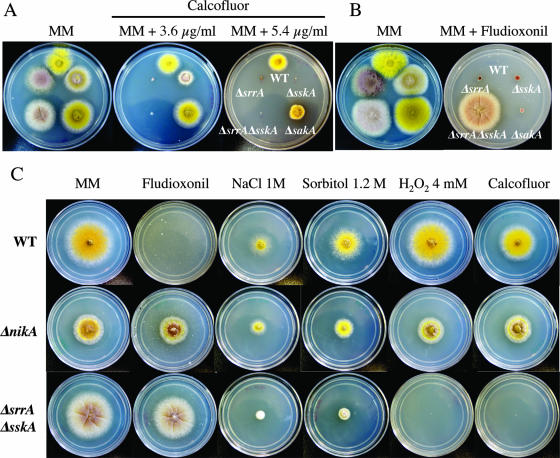
FIG. 4.
RRs SrrA and SskA, the MAPK SakA, and the HK NikA play differential roles in cell wall integrity, fungicide sensitivity, and resistance to osmotic and oxidative stress. Strains CLK43 (wild type [WT]), COSΔsrrA03 (ΔsrrA), COSΔsskA02 (ΔsskA), COSΔsrrA/ΔsskA02 (ΔsrrA ΔsskA), TOL1 (ΔsakA), and CIVΔnikA3 (ΔnikA) were point inoculated onto supplemented MM plates containing either 3.6 to 5.4 μg/ml of the cell wall-disturbing agent calcofluor (A), 2 μg/ml of the fungicide fludioxonil (B and C), or the indicated amount of NaCl, sorbitol, or H2O2 or 5.4 μg/ml of calcofluor (C) and were incubated at 37°C for 3 days. Strains shown contain the yA2 allele, which confers a yellow-spore phenotype; see Table 2 for full strain genotypes.
ΔsskA and ΔsrrA ΔsskA mutants are more sensitive to osmotic stress than ΔsrrA mutants.
To first dissect the roles of SrrA and SskA in stress signal transduction, we tested the growth responses of ΔsrrA, ΔsskA, and ΔsrrA ΔsskA mutants in the presence of 1.2 M KCl (data not shown), 0.6 M NaCl, or 1.2 M sorbitol (Fig. 2). Compared to the wild-type strain, the ΔsrrA mutant showed a slight reduction in radial growth under these conditions. In contrast, growth reduction was more evident for the ΔsskA mutant, particularly in 1.2 M sorbitol, and was even more pronounced for the ΔsrrA ΔsskA mutant (Fig. 2). Thus, although both RRs are required for full resistance to high-osmolarity stress, SskA plays a more prominent role in this response. Moreover, it was noticed that under moderately high osmolarity (0.6 to 0.8 M NaCl or sorbitol), the ΔsrrA mutants not only grew better than the ΔsskA mutants, but asexual sporulation was also improved under these conditions (Fig. 2; see below). These results suggest that although activation of SskA by osmotic stress might compensate for the lack of SrrA, both RRs play significant roles in the process of conidiation.
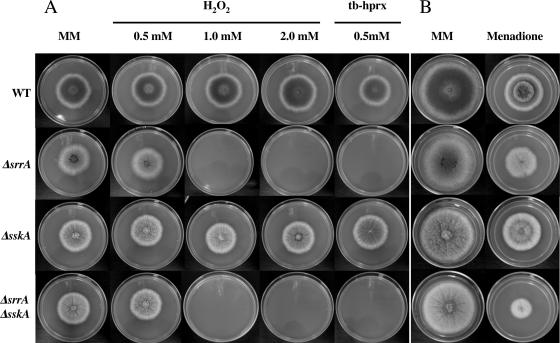
SrrA is required to survive hyperoxidant stress conditions.
Next, we tested the growth responses of ΔsrrA, ΔsskA, and ΔsrrA ΔsskA mutants on plates containing either H2O2, tert-butyl hydroperoxide (tb-hprx), or the superoxide-generating compound menadione (23). We found that while ΔsskA mutants were able to resist up to 2 mM H2O2 and 0.5 mM tb-hprx, ΔsrrA and ΔsrrA ΔsskA double mutants were not (Fig. 3A). In fact, ΔsskA mutants were capable of growing well in the presence of 4 mM H2O2 (data not shown). In the presence of 60 μM menadione, the growth of all strains was moderately reduced. However, ΔsrrA and ΔsrrA ΔsskA mutants were more sensitive to this compound (Fig. 3B). These results indicate that the function of SrrA is essential for the cells to cope with the oxidative stress caused by H2O2 and, to a lesser extent, that provoked by menadione.
FIG. 3.
SrrA is required for resistance to oxidative stress. (A) Strains 11035 (wild type [WT]), TΔsrrA-pyrG7 (ΔsrrA), TΔsskA-riboB8 (ΔsskA), and TΔsrrA-pyrG/ΔsskA-riboB8 (ΔsrrA ΔsskA) were point inoculated onto supplemented MM plates containing either 0.5, 1.0, or 2.0 mM H2O2 or 0.5 mM tb-hprx and were incubated at 37°C for 5 days. (B) The same strains were inoculated as for panel A onto plates containing 60 μM menadione.
ΔsrrA and ΔsskA mutants are sensitive to cell wall stress.
As mentioned earlier, ΔsrrA colonies show irregular borders, and when observed under the microscope, hyphae often show irregular shapes (i.e., not smooth and cylindrical). This and the fact that the SrrA ortholog Skn7 regulates cell wall biosynthesis in S. cerevisiae (42) prompted us to test the sensitivities of RR mutants to the cell wall-disturbing agent calcofluor. We included a ΔsakA mutant in this test, because SakA is part of the SskA pathway and because its ortholog Hog1 has been shown to be needed for calcofluor action (20). The results in Fig. 4A show that the ΔsrrA and ΔsrrA ΔsskA mutants were more sensitive to calcofluor than the ΔsskA mutant. At a higher calcofluor concentration (5.4 μg/ml), the growth of the ΔsskA mutant was completely inhibited, whereas the ΔsakA mutant showed only moderate sensitivity compared to that of the wild type. These results suggest that although both RRs are involved in maintaining cell wall integrity under normal osmolarity, SrrA plays a major role in this process.
SrrA and SskA mediate fungicide sensitivity downstream of a HAMP (histidine kinase, adenylyl cyclase, methyl-binding protein, phosphatase) domain histidine kinase.
It has been shown that fungicides such as fludioxonil activate the osmosensitive MAPK pathway in filamentous fungi and that strains with null mutations in this pathway are resistant to fungicides (36, 77). The A. nidulans MAPK SakA is activated by high osmolarity and oxidative stress (34) in a process mediated by SskA (17). Therefore, to dissect the roles of SrrA, SskA, and SakA in fungicide sensitivity, we tested the growth of these mutants in the presence of fludioxonil. Although single ΔsrrA, ΔsskA, and ΔsakA mutants were slightly less sensitive to fludioxonil than the wild-type strain, only the double ΔsrrA ΔsskA mutant was fully resistant to the fungicide concentration tested. Thus, in A. nidulans, the RR SrrA mediates fludioxonil sensitivity independently of the SskA-SakA pathway (Fig. 4B).
While this paper was in preparation, Izumitsu et al. (30) reported that RRs ChSsk1 and ChSkn7 in Cochliobolus heterostrophus control high-osmolarity adaptation and fungicide sensitivity, under the regulation of the histidine kinase Dic1. This led us to delete the orthologous HK (see Fig. S3B in the supplemental material) encoded by the AN4479.3 gene (19) and to evaluate its role in SrrA and SskA functions. Like Dic1, AN4479.3 encodes an HK with multiple HAMP domains (6) and belongs to class III HKs according to Catlett et al. (14). During the course of this work, Hagiwara et al. (22) published a report in which they named the AN4479.3 gene nikA and analyzed exclusively its role in fungicide resistance. In consequence, we use the name NikA when referring to the HK encoded by AN4479.3. When transformant TΔnikA-pyrG(ΔnkuA)9 was crossed with strain CLK43 (Table 2; see also Fig. S3B in the supplemental material), we observed Mendelian segregation of the ΔnikA mutant phenotype (see below). From the same cross, we obtained the ΔnikA allele in a nkuA+ background.
As shown in Fig. 4C, mutants lacking NikA show decreased radial growth compared to that of wild-type and ΔsrrA ΔsskA strains (∼40% reduction in colony diameter after 3 days of growth). Notably, ΔnikA conidiospores became darkly pigmented with time, forming a distinctive circle in the center of the colony. As in the case of ΔsrrA ΔsskA mutants, the growth of ΔnikA mutants was barely affected by the concentration of fludioxonil tested. These results confirm those of Izumitsu et al. (30) and Hagiwara et al. (22), indicating that in different (but not all) fungi, a HAMP domain HK is required to relay fungicide signals to RRs such as SrrA and SskA.
In light of these results, it became important to analyze the responses of ΔnikA mutants to high osmolarity, oxidative stress, and cell wall stress conditions. Somewhat unexpectedly, ΔnikA mutants were not hypersensitive to high osmolarity. Accordingly, a similar reduction in colony diameter was observed when the wild-type and ΔnikA strains were grown under high osmolarity. Furthermore, the ΔnikA mutant grew better than the ΔsrrA ΔsskA mutant under this condition (Fig. 4C). This indicates the involvement of NikA-independent pathways in high-osmolarity adaptation in A. nidulans. The results in Fig. 4C also show that in contrast to the ΔsrrA and double ΔsrrA ΔsskA mutants, ΔnikA mutants were resistant to 4 mM H2O2, as well as to 0.5 mM tb-hprx (data not shown). Likewise, in contrast to ΔsrrA, ΔsskA, and ΔsrrA ΔsskA mutants, all of which were sensitive to 5.4 μg/ml of calcofluor (Fig. 4A), ΔnikA mutants were able to grow at this calcofluor concentration. This suggests that NikA does not act upstream of SrrA in response to oxidative stress signals and that this HK is not required for SrrA and SskA functions in the biosynthesis of putative cell wall components needed for calcofluor resistance.
Taken together, our results indicate that NikA is required to transmit fungicide signals to the RRs SrrA and SskA but is partially dispensable for transmission of osmotic and oxidative stress signals.
SrrA, SskA, and NikA are required for normal asexual sporulation and conidiospore viability.
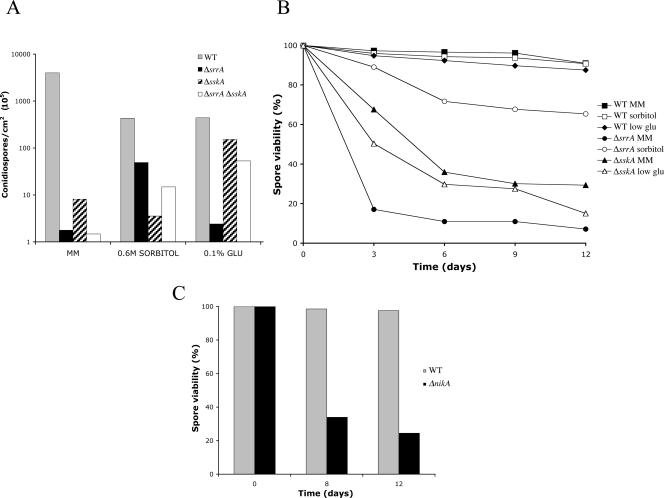
As seen in Fig. 2 to 4, sporulating colonies from ΔsrrA and ΔsskA mutants show reductions in conidiophore density and conidiospore pigmentation. In addition, we found that as with ΔsakA mutants (34), ΔsrrA and ΔsskA conidiospores lost their viability during storage. These observations led us to examine in more detail the conidiation process, as well as conidiospore viability, for these mutants. Since it was evident that ΔsrrA and ΔsskA sporulation defects were partially suppressed under high-osmolarity and low-glucose conditions, respectively (see Fig. S4 in the supplemental material), we decided to quantify the number of spores formed under these conditions. The results in Fig. 5A show that compared to the wild-type strain, ΔsrrA and ΔsskA mutants produce very low numbers of spores in MM. This sporulation defect was slightly more severe in ΔsrrA and ΔsrrA ΔsskA mutants. Consistent with our previous observations, we found that sporulation of the ΔsrrA but not the ΔsskA mutant was greatly improved at high sorbitol concentrations. In contrast, ΔsskA but not ΔsrrA sporulation was notably improved at low glucose concentrations. These results demonstrate that both RRs regulate sporulation in A. nidulans, and they suggest that SskA stimulation by high osmolarity results in increased sporulation, while low glucose stimulates sporulation in a SrrA-dependent fashion. Unexpectedly, the sporulation of the ΔsrrA ΔsskA double mutant was improved under both high-sorbitol and low-glucose conditions (Fig. 5A), suggesting that in the absence of both RRs, other sporulation pathways might become activated.
FIG. 5.
SrrA, SskA, and NikA are required for asexual sporulation and conidiospore viability. (A) Strains 11035 (wild type [WT]), TΔsrrA-pyrG7 (ΔsrrA), TΔsskA-riboB8 (ΔsskA), and TΔsrrA-pyrG/ΔsskA-riboB8 (ΔsrrA ΔsskA) were point inoculated onto supplemented MM plates or onto plates containing MM with 0.6 M sorbitol or MM with low (0.1%) glucose concentrations and were incubated at 37°C for 6 days. Total conidiospores per colony were harvested, counted, and the count divided by the colony area to obtain the number of conidiospores per square centimeter. Conidial yield data are means for two independent colonies. (B) Conidiospore suspensions from the experiment for which results are shown in panel A were plated immediately after counting or maintained at 4°C for up to 12 days. At the indicated time points, aliquots were diluted appropriately and used to inoculate supplemented MM plates, which were incubated at 37°C for 2 days; then colonies were counted. (C) Strains CLK43 (WT) and CIVΔnikA3 (ΔnikA) were point inoculated onto supplemented MM plates and incubated at 37°C for 6 days. Conidia were collected, counted, divided by the colony area, and treated as for panel B. For panels B and C, the number of germinated spores at the time of harvesting was considered as 100% viability for each strain. The number of colonies counted ranged from 127 to 214 per plate. Data shown are means for two independent plates, showing a maximum variation of 17% with respect to the mean.
In light of these results, we decided to test not only the viability of the spores produced in MM but also the viability of those produced under high-osmolarity and low-glucose conditions. For this purpose, spores were stored as a water suspension at 4°C, a condition often used for short-term storage of A. nidulans spores. The results in Fig. 5B show that while the viability of wild-type spores remained virtually unaffected after 12 days, the viability of ΔsrrA and ΔsskA spores was drastically reduced after only 3 to 6 days at 4°C. Furthermore, it was found that the viability of ΔsrrA spores produced under high-sorbitol conditions was greatly improved. In contrast, the viability of ΔsskA spores produced under low-glucose conditions remained low. These results indicate that both RRs are required for full sporulation as well as for full spore viability and that SskA might replace SrrA functions in these two processes.
After we found that the HK NikA acts upstream of SrrA and SskA in fungicide signaling, it was interesting to evaluate its role in conidiation and conidiospore viability. We found that the ΔnikA mutant produced ∼30 times fewer conidiospores per area than the wild-type strain. In comparison, ΔsskA mutants produced 500 times fewer conidia than the wild type (Fig. 5A), and thus, the ΔnikA conidiation defect was not as marked as that observed for the ΔsrrA or ΔsskA mutant. In contrast, ΔnikA conidiospores showed a decrease in viability that was similar to that observed for ΔsskA spores (Fig. 5B and C). These results suggest that, acting through SskA or SrrA, NikA and other HKs appear to contribute to the sporulation process. In contrast, NikA appears to play a major role in regulating spore viability, possibly through the RR SskA.
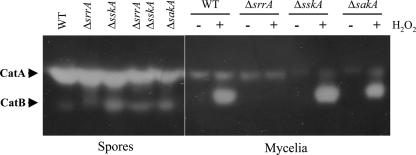
SrrA regulates expression of the catalase gene catB.
We reported previously that the viability of ΔsakA conidiospores decreased upon storage and that the activity of the spore-specific catalase CatA (48) was lower in the ΔsakA mutant than in the wild type (34). Since SskA acts upstream of SakA and since ΔsskA and ΔsrrA conidiospores show decreased viability (Fig. 5B), we analyzed catalase activity levels in these mutants to determine if there was a correlation between catalase activity, spore viability, and H2O2 resistance. The results show that the CatA activity levels of ΔsrrA and ΔsskA conidiospores were comparable to those found for wild-type spores. CatA activity levels of ΔsrrA ΔsskA and ΔsakA conidiospores were lower (Fig. 6, left panel). These results indicate that SrrA and SskA are individually dispensable for CatA accumulation in conidia, and thus, CatA activity cannot explain the observed decreases in viability or H2O2 resistance of ΔsrrA and ΔsskA mutants. However, for A. nidulans, catalase CatB constitutes the major inducible catalase activity in mycelia (33, 35). Therefore, we examined SrrA and SskA functions in CatB induction. The results show that H2O2 induces high levels of CatB activity in both the wild-type strain and the ΔsskA mutant. In contrast, no induction of CatB activity was observed for the ΔsrrA mutant, indicating a critical role of SrrA in CatB induction by H2O2. In addition, it was noted that CatB induction was lower in the ΔsakA mutant than in the wild-type strain. This is consistent with previous results and indicates that CatB induction seems partially dependent on the MAPK SakA, which in turn suggests some interaction between SrrA and SakA in catB transcriptional regulation (Fig. 6, right panel). Taken together, our results indicate that the H2O2 sensitivity of ΔsrrA mutants is related to their failure to induce CatB. Consistent with this, ΔsskA mutants are able to resist H2O2 (Fig. 3) and to induce CatB (Fig. 6).
FIG. 6.
Catalase activities in conidiospores and H2O2-treated mycelia from ΔsrrA, ΔsskA, and ΔsakA mutants. Protein extracts were prepared from conidiospores (left panel) or H2O2-treated mycelia (right panel) from strains CLK43 (wild type [WT]), COSΔsrrA03 (ΔsrrA), COSΔsskA02 (ΔsskA), COSΔsrrA/ΔsskA02 (ΔsrrA ΔsskA), and TOL1 (ΔsakA). Thirty micrograms of total protein was separated on native polyacrylamide gels to determine catalase activity as described previously (35, 48). Conidiospores were harvested from 6-day colonies. Mycelia grown for 8 h were treated with 0.5 mM H2O2 as described previously (35).
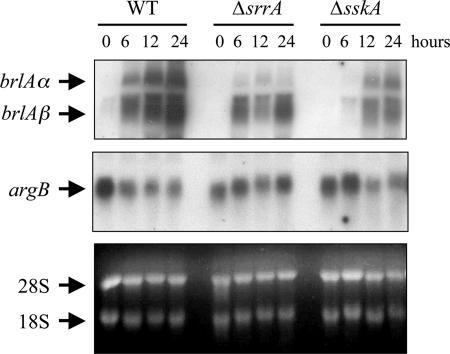
SrrA and SskA regulate brlA mRNA accumulation.
Asexual reproduction in A. nidulans depends on the activity of the transcription factor BrlA (1, 16). Furthermore, the brlA mRNA levels have been correlated with conidiophore morphology and conidiospore production (3, 13, 60, 66). Therefore, we examined brlA mRNA levels in ΔsrrA and ΔsskA mutants that were induced to conidiate. As reported previously, the two brlA transcripts (60) are not present before induction of conidiation and then gradually accumulate during wild-type conidiation. In the ΔsrrA mutant, brlA mRNA started to accumulate at the same time but then failed to reach wild-type levels. A similar result was observed for the ΔsskA mutant, except that in this case brlA mRNA accumulation is delayed by 6 h relative to that for the wild type (Fig. 7). As in other cases (66), brlA mRNA appears somewhat diffused. However, probing of the same blot with the argB gene, whose expression remains relatively constant during conidiation, indicates that this is not due to general RNA degradation. In any case, brlA mRNA accumulation levels appear decreased in ΔsrrA and ΔsskA mutants, suggesting that this might be related to the sporulation defects observed for these mutants.
FIG. 7.
Accumulation of brlA mRNA during conidiation is reduced in ΔsrrA and ΔsskA mutants. Liquid cultures from strains CLK43 (wild type [WT]), COSΔsrrA03 (ΔsrrA), and COSΔsskA02 (ΔsskA) were grown for 18 h (0 h of conidiation) and induced to conidiate. Total-RNA blots from samples taken at the indicated times were hybridized with brlA- and argB-specific probes. The positions of brlA and argB transcripts and the rRNAs are indicated. The bottom panel shows rRNA as a loading reference. For all strains, 0 h of development (18 h of growth) corresponds to growing hyphae. WT development corresponds to young conidiophores without conidia (6 h), conidiophores and immature conidia (12 h), and mature conidiophores and conidia (24 h).
DISCUSSION
In A. nidulans, the deletion of the HPt protein YpdA results in lethality.
We used a genetic approach to analyze a phosphorelay system involved in stress signaling and asexual development in A. nidulans. This analysis started with the cloning and characterization of the srrA gene as a new member of the skn7/prr1 family and was extended to analyze how SrrA was related to SskA, a second RR, as well as to the HPt protein YpdA and the HK NikA as upstream components of this system. We were able to delete the ypdA gene and show that the ΔypdA allele could be maintained only in heterokaryons. Notably, the growth of ypdA−/ypdA+ heterokaryons, which appear to contain a higher proportion of ypdA+ nuclei, was reduced or eliminated under osmotic but not oxidative stress conditions (Fig. 1A to D). Although the interpretation of these results is complicated by the inherent genetic heterogeneity of heterokaryons, these results suggest that YpdA plays a more important role in osmoadaptation than in adaptation to oxidative stress. The fact that mutants lacking YpdA are very likely unviable contrasts with the fact that mutants lacking both RRs (ΔsrrA ΔsskA) are viable, although clearly affected in stress signal transduction and asexual sporulation. This argues against a model in which YpdA simply exerts positive effects over both RRs and these in turn play positive roles for downstream components. By analogy with S. cerevisiae, in which ypd1 mutation results in lethal hyperactivation of the Hog1 MAPK (58), we propose that the lethality associated with YpdA elimination results from hyperactivation of SakA and/or MpkC, a second stress MAPK present in the aspergilli (17, 34), whose inactivation does not produce a clear phenotype in A. nidulans (K. Jahng, personal communication) (17). However, MpkC functionality is suggested by the facts that in Aspergillus fumigatus, mpkC deletion results in the inability to use polyalcohol sugars as a sole carbon source and that mpkC mRNA levels increase during oxidative stress (61). That constitutive activation of SakA and/or MpkC can lead to lethality is also indicated by the fact that the fungicide fludioxonil seems to kill A. nidulans (Fig. 4B and C) and other fungi through activation of the stress MAPK pathway (36, 77) and, as shown here, through the activity of the RR SrrA.
SrrA and SskA roles in osmotic and fungicide stress signal transduction.
We found that both SskA and SrrA are involved in osmotic stress signal transduction. Although the growth of the wild type and ΔsrrA mutants is similar under moderately high osmolarity (0.6 M sorbitol or KCl), the role of SrrA in osmotic stress resistance is inferred from the fact that ΔsrrA ΔsskA double mutants are more sensitive to osmostress than single ΔsskA mutants. On the other hand, the fact that ΔsskA mutants are more sensitive to osmotic stress than ΔsakA mutants (34) suggests that SskA activates SakA and MpkC and that both MAPKs might contribute to osmostress resistance. This is supported by the fact that PbsB, a MAPK kinase acting downstream of SskA, is required for phosphorylation of both SakA and MpkC (17). Testing of this simple hypothesis has been prevented by an apparent lethality of double ΔsakA ΔmpkC mutants (4, 17). If confirmed, this hypothesis will imply that A. nidulans uses two MAPKs as well as the RR SrrA to deal with high-osmolarity stress.
A finding in this work that might have practical consequences in terms of fungicide resistance mechanisms is the fact that both SrrA and SskA are required for sensitivity to the widely used fungicide fludioxonil. There is strong evidence showing that the osmosensing MAPK kinase pathway mediates the killing effects of this broad-spectrum fungicide. Indeed, Neurospora crassa mutants with null mutations of the MAPK OS-2 (77), the RR RRG-1 (31), or the HK OS-1 (52) are both osmosensitive and resistant to fludioxonil. This is in contrast to our results, which show that a lack of SakA, the OS-2 ortholog, does not result in complete resistance to fludioxonil and that, in fact, the RR SrrA mediates fludioxonil sensitivity independently of the SskA-SakA pathway (Fig. 4). We found that like os-2 mutants, ΔnikA mutants are resistant to fludioxonil. This indicates that out of the 15 predicted histidine kinases in A. nidulans, NikA plays a major role in relaying fungicide stress signals to SrrA and SskA. A similar fungicide sensitivity function has recently been found for NikA orthologs BOS1 (72) and Dic1 (30). However, some of these HK orthologs seem to have species-specific functions. For example, BOS1 inactivation in Botrytis cinerea results in lack of macroconidiation and plant virulence (72). In A. nidulans, ΔnikA but not ΔsrrA ΔsskA mutants show a clear reduction in radial growth, suggesting that NikA serves functions that are not mediated by SrrA and/or SskA. In addition, NikA is involved in conidiation and is required for spore viability (Fig. 5C).
We also found that the role of NikA in the osmoadaptation of A. nidulans does not appear as important as in other fungi such as N. crassa. While os-2-null mutants are hypersensitive to osmostress, ΔnikA mutants show partial or no sensitivity to this stress. The fact that ΔsrrA ΔsskA mutants are more sensitive to osmostress than ΔnikA mutants suggests that another HK(s) is able to transmit osmostress signals to SrrA and/or SskA in A. nidulans. TcsB, the ortholog of the yeast HK Sln1, is a likely candidate to mediate osmostress signals in different fungi. Although TcsB inactivation did not produce a visible phenotype (18), it would be interesting to evaluate the osmostress response of double ΔnikA ΔtcsB mutants.
SrrA, SskA, and NikA roles in oxidative stress signal transduction and calcofluor sensitivity.
We found that SrrA but not SskA is required for H2O2 resistance and, consistent with this, for the induction of catalase CatB in response to H2O2. This is coherent with the roles that the SrrA orthologs Skn7/Pos1 and Prr1 play in antioxidant responses in S. cerevisiae and S. pombe, respectively. However, the disruption of skn7 in Cryptococcus neoformans did not affect H2O2 sensitivity (7), suggesting that the antioxidant response through this RR might not be conserved in all fungi. Notably, the Skn7 function in oxidative stress response does not require the conserved phosphoaccepting aspartate present in its receiver domain (41, 46). The fact that ΔnikA mutants are resistant to H2O2 indicates that either another HK(s) is responsible for transmitting oxidative stress signals to SrrA or the SrrA function in the antioxidant response does not involve phosphorelay components. Likewise, the resistance of ΔnikA mutants to calcofluor suggests that another HK(s) can regulate SrrA and/or SskA functions in cell wall integrity.
Since SskA is required for transmission of oxidative stress signals to the MAPK SakA, it was unexpected that ΔsskA mutants were not sensitive to H2O2 and were able to induce CatB in response to H2O2. However, a role for the SskA-SakA pathway in the oxidative stress response cannot be ruled out. In fact, H2O2-mediated induction of a catB::lacZ reporter gene fusion is reduced in a ΔsakA background (32), and CatA activity levels are reduced in ΔsakA conidiospores (34) (Fig. 6).
The roles of SrrA, SskA, and NikA in asexual development and conidiospore viability.
We have shown that two RRs and one HK differentially contribute to the development of asexual spores as well as to the viability of these spores. Indeed, the inactivation of srrA, sskA, or nikA led to the production of fewer spores that showed decreased viability upon storage. Two observations indicate that, although related, spore formation and spore viability are separable processes. First, ΔsakA and wild-type strains produce similar amounts of spores, but only the ΔsakA spores lose their viability (34). Second, the sporulation of ΔsskA mutants is greatly improved under low-glucose conditions, but the spores produced maintain their decreased viability. In contrast, under high-osmolarity conditions, ΔsrrA mutants clearly improve both sporulation and conidiospore viability (Fig. 5A and B). The fact that ΔsrrA and ΔsskA mutants are sensitive to calcofluor (Fig. 4A) suggests that their cell wall biosynthesis is affected, which in turn could be related to cell wall-defective spores and decreased viability. However, ΔsakA mutants are rather resistant to calcofluor (Fig. 4A), yet ΔsakA spores show decreased viability (34).
The results showing that ΔsrrA and ΔsskA sporulation defects are clearly more drastic than those seen in ΔnikA mutants suggest that SrrA and/or SskA functions in sporulation might be regulated by other upstream HKs. This is supported by the facts that inactivation of the HK TcsA resulted in reduced conidiation and that this defect was remedied under high osmolarity (73).
While we do not know why spore viability decreases in mutants lacking SakA, SrrA, SskA, or NikA, the decreased sporulation observed for ΔsrrA and ΔsskA mutants might be related to their lower levels of brlA mRNA during sporulation (Fig. 7). BrlA is a rate-limiting step function during conidiospore formation, required not only for initiation of conidiation but throughout the entire process. Because of the close link between decreased brlA mRNA levels and decreased sporulation, it is possible that SrrA and SskA functions affect brlA mRNA accumulation and that this in turn affects the initiation and/or maintenance of the conidiation process. Reduced sporulation has been related to reduced brlA expression in several “fluffy” developmental mutants (2, 39, 65, 67, 74). However, ΔsrrA and ΔsskA mutants might represent a different class of sporulation mutants. First, ΔsrrA and ΔsskA mutants do not show a “fluffy” phenotype, characterized by the proliferation of aerial hyphae. Second, ΔsrrA and ΔsskA mutants are able to induce conidiation of ΔfluG and ΔtmpA “fluffy” mutants when grown next to them (data not shown). This suggests that ΔsrrA and ΔsskA mutants are not defective in the production of the putative sporulation signals produced by FluG (39, 65) and TmpA (67).
Mutants with null mutations of the devR gene show conidiation defects similar to those found for ΔsrrA mutants, and these defects are remedied by high osmolarity (71). devR encodes a helix-loop-helix transcription factor proposed to be a downstream target of the phosphorelay system. This is because a DevR-green fluorescent protein fusion failed to show nuclear localization in a mutant in which the HK gene tcsA was affected (71, 73). We found that devR mutants are not as sensitive to H2O2 as ΔsrrA mutants (data not shown), suggesting that if DevR acts downstream of SrrA, it mediates sporulation functions. The role of DevR in conidiospore viability has not been evaluated. How conserved the SrrA function in regulating sporulation is among fungi has yet to be determined. Recently, it was reported that A. fumigatus mutants lacking the orthologous RR AfSkn7 are sensitive to oxidative stress but do not show growth or conidiation phenotypes, even under high-osmolarity conditions (38).
The function of SskA in spore viability is consistent with its function as an upstream component of the SakA pathway (17). Indeed, SakA is phosphorylated shortly after induction of asexual development, and its inactivation results in spores with decreased viability (34). The fact that ΔsakA conidia contained lower levels of the spore-specific catalase CatA suggested a relationship between CatA content and spore viability. However, we found here that ΔsrrA, ΔsskA, and wild-type spores contain comparable levels of CatA activity.
While this work was under review, Hagiwara and coworkers (21) reported the characterization of ΔsrrA, ΔsskA, and ΔsrrA ΔsskA mutants with regard to their resistance to osmotic and oxidative stress, conidiospore viability, and the mRNA accumulation of several genes, including catalase genes. Some of the results reported are consistent with ours, but others show important differences. First, Hagiwara and coworkers report that both ΔsrrA and ΔsskA mutants are sensitive to H2O2; we find that only ΔsrrA mutants are sensitive to H2O2, within a 1 to 4 mM range. Second, the cited work shows that the srrA and sskA genes are both required for catB gene expression. By directly measuring catalase activity, we find that only srrA is fully required for CatB induction. Third, the report of Hagiwara et al. shows that only ΔsskA conidiospores lose viability; we show that both ΔsrrA and ΔsskA conidiospores show decreased viability. Fourth, Hagiwara and colleagues report that catA gene expression is downregulated in ΔsrrA and ΔsskA conidiospores. By directly measuring catalase activity, we find that neither ΔsrrA nor ΔsskA conidiospores show lower CatA activity. It seems unlikely that these differences are due to strain variations, because we have confirmed our results using strains with different genetic backgrounds. Although currently we do not have an explanation for these discrepancies, it will be important to consider them in designing future experiments.
Our results highlight important differences in the ways different fungi perceive and respond to stress signals and in the ways in which phosphorelay systems are connected to developmental processes. Taken together, these results support a model in which the A. nidulans HK NikA transmits fungicide stress signaling to the SskA-SakA pathway, as well as to the RR SrrA, resulting in cell death. NikA might also transmit osmostress signals to SrrA and/or SskA, but other HKs seem important for osmoadaptation. We propose that NikA is also involved in transmitting unknown signals to contribute to asexual sporulation and to determine conidiospore viability, probably via SskA. In addition, NikA appears to have SrrA/SskA-independent functions in the regulation of radial growth.
In our model, SrrA plays a major role in the oxidative stress response independently of NikA and perhaps independently of any other phosphorelay component. Both RRs are required for normal asexual sporulation and for maintenance of conidiospore viability, perhaps through regulation of brlA gene mRNA levels. Finally, SrrA and SskA are very likely involved in cell wall biosynthesis.
Acknowledgments
This work was supported by grants 2002-C01-40026, CB-2005-01-49667, and 47799-Q from CONACYT and by grants IN228507 and IN221106/17 from PAPIIT-UNAM (México). I. Vargas-Pérez was supported by a scholarship from CONACYT.
We are grateful to Anet Rivera for experimental support, to Fernando Lara for nkuA primer design, and to the IFC-UNAM molecular biology unit for DNA synthesis and sequencing. We thank Michael Hynes and Reinhard Fischer for providing very useful strains.
Footnotes
Published ahead of print on 13 July 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adams, T. H., M. T. Boylan, and W. E. Timberlake. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353-362. [DOI] [PubMed] [Google Scholar]
- 2.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguirre, J. 1993. Spatial and temporal controls of the Aspergillus brlA developmental regulatory gene. Mol. Microbiol. 8:211-218. [DOI] [PubMed] [Google Scholar]
- 4.Aguirre, J., W. Hansberg, and R. Navarro. 2006. Fungal responses to reactive oxygen species. Med. Mycol. 44:S101-S107. [DOI] [PubMed] [Google Scholar]
- 5.Aguirre, J., M. Rios-Momberg, D. Hewitt, and W. Hansberg. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13:111-118. [DOI] [PubMed] [Google Scholar]
- 6.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 7.Bahn, Y. S., K. Kojima, G. M. Cox, and J. Heitman. 2006. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 17:3122-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumenstein, A., K. Vienken, R. Tasler, J. Purschwitz, D. Veith, N. Frankenberg-Dinkel, and R. Fischer. 2005. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr. Biol. 15:1833-1838. [DOI] [PubMed] [Google Scholar]
- 9.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. Plamann, M. Goodrich-Tanrikulu, U. Schulte, G. Mannhaupt, F. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Sachs, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, J. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brody, H., J. Griffith, A. J. Cuticchia, J. Arnold, and W. E. Timberlake. 1991. Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res. 19:3105-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, J. L., H. Bussey, and R. C. Stewart. 1994. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 13:5186-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck, V., J. Quinn, T. Soto Pino, H. Martin, J. Saldanha, K. Makino, B. A. Morgan, and J. B. Millar. 2001. Peroxide sensors for the fission yeast stress-activated mitogen-activated protein kinase pathway. Mol. Biol. Cell 12:407-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busby, T. M., K. Y. Miller, and B. L. Miller. 1996. Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of the bristle and stunted genes. Genetics 143:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catlett, N. L., O. C. Yoder, and B. G. Turgeon. 2003. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell 2:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chauhan, N., J. P. Latge, and R. Calderone. 2006. Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Rev. Microbiol. 4:435-444. [DOI] [PubMed] [Google Scholar]
- 16.Clutterbuck, A. J. 1969. A mutational analysis of conidial development in Aspergillus nidulans. Genetics 63:317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furukawa, K., Y. Hoshi, T. Maeda, T. Nakajima, and K. Abe. 2005. Aspergillus nidulans HOG pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol. Microbiol. 56:1246-1261. [DOI] [PubMed] [Google Scholar]
- 18.Furukawa, K., Y. Katsuno, T. Urao, T. Yabe, T. Yamada-Okabe, H. Yamada-Okabe, Y. Yamagata, K. Abe, and T. Nakajima. 2002. Isolation and functional analysis of a gene, tcsB, encoding a transmembrane hybrid-type histidine kinase from Aspergillus nidulans. Appl. Environ. Microbiol. 68:5304-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 20.García-Rodriguez, L. J., A. Duran, and C. Roncero. 2000. Calcofluor antifungal action depends on chitin and a functional high-osmolarity glycerol response (HOG) pathway: evidence for a physiological role of the Saccharomyces cerevisiae HOG pathway under noninducing conditions. J. Bacteriol. 182:2428-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagiwara, D., Y. Asano, J. Marui, K. Furukawa, K. Kanamaru, M. Kato, K. Abe, T. Kobayashi, T. Yamashino, and T. Mizuno. 2007. The SskA and SrrA response regulators are implicated in oxidative stress responses of hyphae and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71:1003-1014. [DOI] [PubMed] [Google Scholar]
- 22.Hagiwara, D., Y. Matsubayashi, J. Marui, K. Furukawa, T. Yamashino, K. Kanamaru, M. Kato, K. Abe, T. Kobayashi, and T. Mizuno. 2007. Characterization of the NikA histidine kinase implicated in the phosphorelay signal transduction of Aspergillus nidulans, with special reference to fungicide responses. Biosci. Biotechnol. Biochem. 71:844-847. [DOI] [PubMed] [Google Scholar]
- 23.Halliwell, B., and J. M. C. Gutteridge. 1989. Free radicals in biology and medicine, 2nd ed. Clarendon Press, Oxford, United Kingdom.
- 24.Han, K. H., and R. A. Prade. 2002. Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol. Microbiol. 43:1065-1078. [DOI] [PubMed] [Google Scholar]
- 25.Hansberg, W., and J. Aguirre. 1990. Hyperoxidant states cause microbial cell differentiation by cell isolation from dioxygen. J. Theor. Biol. 142:201-221. [DOI] [PubMed] [Google Scholar]
- 26.Hill, T. W., and E. Käfer. 2001. Improved protocols for Aspergillus minimal medium: trace element and minimal medium stock solution. Fungal Genet. Newsl. 48:20-21. [Google Scholar]
- 27.Hoch, J. A., and T. J. Silhavy (ed.). 1995. Two-component signal transduction. ASM Press, Washington, DC.
- 28.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikner, A., and K. Shiozaki. 2005. Yeast signaling pathways in the oxidative stress response. Mutat. Res. 569:13-27. [DOI] [PubMed] [Google Scholar]
- 30.Izumitsu, K., A. Yoshimi, and C. Tanaka. 2007. Two-component response regulators Ssk1p and Skn7p additively regulate high-osmolarity adaptation and fungicide sensitivity in Cochliobolus heterostrophus. Eukaryot. Cell 6:171-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, C. A., S. E. Greer-Phillips, and K. A. Borkovich. 2007. The response regulator RRG-1 functions upstream of a mitogen-activated protein kinase pathway impacting asexual development, female fertility, osmotic stress, and fungicide resistance in Neurospora crassa. Mol. Biol. Cell 18:2123-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawasaki, L. 1988. Las catalasas y la respuesta antioxidante durante el crecimiento y la diferenciación en Aspergillus nidulans. Ph.D. thesis. Universidad Nacional Autonoma de Mexico, D.F., Mexico.
- 33.Kawasaki, L., and J. Aguirre. 2001. Multiple catalase genes are differentially regulated in Aspergillus nidulans. J. Bacteriol. 183:1434-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawasaki, L., O. Sanchez, K. Shiozaki, and J. Aguirre. 2002. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45:1153-1163. [DOI] [PubMed] [Google Scholar]
- 35.Kawasaki, L., D. Wysong, R. Diamond, and J. Aguirre. 1997. Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress. J. Bacteriol. 179:3284-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kojima, K., Y. Takano, A. Yoshimi, C. Tanaka, T. Kikuchi, and T. Okuno. 2004. Fungicide activity through activation of a fungal signalling pathway. Mol. Microbiol. 53:1785-1796. [DOI] [PubMed] [Google Scholar]
- 37.Krems, B., C. Charizanis, and K. D. Entian. 1996. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 29:327-334. [DOI] [PubMed] [Google Scholar]
- 38.Lamarre, C., O. Ibrahim-Granet, C. Du, R. Calderone, and J. P. Latgé. 2007. Characterization of the SKN7 ortholog of Aspergillus fumigatus. Fungal Genet. Biol. 44:682-690. [DOI] [PubMed] [Google Scholar]
- 39.Lee, B. N., and T. H. Adams. 1996. fluG and flbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlAβ activation. EMBO J. 15:299-309. [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040-16046. [DOI] [PubMed] [Google Scholar]
- 41.Li, S., A. Ault, C. L. Malone, D. Raitt, S. Dean, L. H. Johnston, R. J. Deschenes, and J. S. Fassler. 1998. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 17:6952-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, S., S. Dean, Z. Li, J. Horecka, R. J. Deschenes, and J. S. Fassler. 2002. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol. Biol. Cell 13:412-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Márquez-Fernández, O., A. Trigos, J. L. Ramos-Balderas, G. Viniegra-Gonzalez, H. B. Deising, and J. Aguirre. 2007. Phosphopantetheinyl transferase CfwA/NpgA is required for Aspergillus nidulans secondary metabolism and asexual development. Eukaryot. Cell 6:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinelli, S. D., and J. R. Kinghorn (ed.). 1994. Progress in industrial microbiology, vol. 29. Aspergillus: 50 years on, 1st ed. Elsevier, Amsterdam, The Netherlands.
- 45.Mohanty, S., S. Lee, N. Yadava, M. J. Dealy, R. S. Johnson, and R. A. Firtel. 2001. Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Dev. 15:1435-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan, B. A., G. R. Banks, W. M. Toone, D. Raitt, S. Kuge, and L. H. Johnston. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgan, B. A., N. Bouquin, G. F. Merrill, and L. H. Johnston. 1995. A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J. 14:5679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navarro, R. E., and J. Aguirre. 1998. Posttranscriptional control mediates cell type-specific localization of catalase A during Aspergillus nidulans development. J. Bacteriol. 180:5733-5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nayak, T., E. Szewczyk, C. E. Oakley, A. Osmani, L. Ukil, S. L. Murray, M. J. Hynes, S. A. Osmani, and B. R. Oakley. 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172:1557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen, A. N., A. Lee, W. Place, and K. Shiozaki. 2000. Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol. Biol. Cell 11:1169-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nixon, B. T., C. W. Ronson, and F. M. Ausubel. 1986. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc. Natl. Acad. Sci. USA 83:7850-7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ochiai, N., M. Fujimura, T. Motoyama, A. Ichiishi, R. Usami, K. Horikoshi, and I. Yamaguchi. 2001. Characterization of mutations in the two-component histidine kinase gene that confer fludioxonil resistance and osmotic sensitivity in the os-1 mutants of Neurospora crassa. Pest Manag. Sci. 57:437-442. [DOI] [PubMed] [Google Scholar]
- 53.Ohmiya, R., C. Kato, H. Yamada, H. Aiba, and T. Mizuno. 1999. A fission yeast gene (prr1+) that encodes a response regulator implicated in oxidative stress response. J. Biochem. (Tokyo) 125:1061-1066. [DOI] [PubMed] [Google Scholar]
- 54.Ohmiya, R., H. Yamada, C. Kato, H. Aiba, and T. Mizuno. 2000. The Prr1 response regulator is essential for transcription of ste11+ and for sexual development in fission yeast. Mol. Gen. Genet. 264:441-451. [DOI] [PubMed] [Google Scholar]
- 55.Ouaked, F., W. Rozhon, D. Lecourieux, and H. Hirt. 2003. A MAPK pathway mediates ethylene signaling in plants. EMBO J. 22:1282-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 57.Posas, F., and H. Saito. 1998. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 17:1385-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865-875. [DOI] [PubMed] [Google Scholar]
- 59.Prade, R. A., P. Ayoubi, S. Krishnan, S. Macwana, and H. Russell. 2001. Accumulation of stress and inducer-dependent plant-cell-wall-degrading enzymes during asexual development in Aspergillus nidulans. Genetics 157:957-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prade, R. A., and W. E. Timberlake. 1993. The Aspergillus nidulans brlA regulatory locus consists of overlapping transcription units that are individually required for conidiophore development. EMBO J. 12:2439-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reyes, G., A. Romans, C. K. Nguyen, and G. S. May. 2006. Novel mitogen-activated protein kinase MpkC of Aspergillus fumigatus is required for utilization of polyalcohol sugars. Eukaryot. Cell 5:1934-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sánchez, O., and J. Aguirre. 1996. Efficient transformation of Aspergillus nidulans by electroporation of germinated conidia. Fungal Genet. Newsl. 43:48-51. [Google Scholar]
- 63.Sánchez, O., R. E. Navarro, and J. Aguirre. 1998. Increased transformation frequency and tagging of developmental genes in Aspergillus nidulans by restriction enzyme-mediated integration (REMI). Mol. Gen. Genet. 258:89-94. [DOI] [PubMed] [Google Scholar]
- 64.Santos, J. L., and K. Shiozaki. 2001. Fungal histidine kinases. Sci. STKE 2001:RE1. [DOI] [PubMed] [Google Scholar]
- 65.Seo, J. A., Y. Guan, and J. H. Yu. 2003. Suppressor mutations bypass the requirement of fluG for asexual sporulation and sterigmatocystin production in Aspergillus nidulans. Genetics 165:1083-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skromne, I., O. Sanchez, and J. Aguirre. 1995. Starvation stress modulates the expression of the Aspergillus nidulans brlA regulatory gene. Microbiology 141:21-28. [DOI] [PubMed] [Google Scholar]
- 67.Soid-Raggi, G., O. Sanchez, and J. Aguirre. 2006. TmpA, a member of a novel family of putative membrane flavoproteins, regulates asexual development in Aspergillus nidulans. Mol. Microbiol. 59:854-869. [DOI] [PubMed] [Google Scholar]
- 68.Thomason, P. A., D. Traynor, J. B. Stock, and R. R. Kay. 1999. The RdeA-RegA system, a eukaryotic phospho-relay controlling cAMP breakdown. J. Biol. Chem. 274:27379-27384. [DOI] [PubMed] [Google Scholar]
- 69.Timberlake, W. E., and A. J. Clutterbuck. 1994. Genetic regulation of conidiation. Prog. Ind. Microbiol. 29:383-427. [PubMed] [Google Scholar]
- 70.Tsitsigiannis, D. I., and N. P. Keller. 2007. Oxylipins as developmental and host-fungal communication signals. Trends Microbiol. 15:109-118. [DOI] [PubMed] [Google Scholar]
- 71.Tüncher, A., H. Reinke, G. Martic, M. L. Caruso, and A. A. Brakhage. 2004. A basic-region helix-loop-helix protein-encoding gene (devR) involved in the development of Aspergillus nidulans. Mol. Microbiol. 52:227-241. [DOI] [PubMed] [Google Scholar]
- 72.Viaud, M., S. Fillinger, W. Liu, J. S. Polepalli, P. Le Pecheur, A. R. Kunduru, P. Leroux, and L. Legendre. 2006. A class III histidine kinase acts as a novel virulence factor in Botrytis cinerea. Mol. Plant-Microbe Interact. 19:1042-1050. [DOI] [PubMed] [Google Scholar]
- 73.Virginia, M., C. L. Appleyard, W. L. McPheat, and M. J. Stark. 2000. A novel ‘two-component’ protein containing histidine kinase and response regulator domains required for sporulation in Aspergillus nidulans. Curr. Genet. 37:364-372. [DOI] [PubMed] [Google Scholar]
- 74.Wieser, J., B. N. Lee, J. Fondon III, and T. H. Adams. 1994. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr. Genet. 27:62-69. [DOI] [PubMed] [Google Scholar]
- 75.Yu, J. H., Z. Hamari, K. H. Han, J. A. Seo, Y. Reyes-Dominguez, and C. Scazzocchio. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973-981. [DOI] [PubMed] [Google Scholar]
- 76.Yu, J. H., J. Wieser, and T. H. Adams. 1996. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15:5184-5190. [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, Y., R. Lamm, C. Pillonel, S. Lam, and J. R. Xu. 2002. Osmoregulation and fungicide resistance: the Neurospora crassa os-2 gene encodes a HOG1 mitogen-activated protein kinase homologue. Appl. Environ. Microbiol. 68:532-538. [DOI] [PMC free article] [PubMed] [Google Scholar]