Abstract
Microarray analysis was used to identify transcriptional changes in early vegetative growth of the filamentous fungus Aspergillus nidulans. The results suggest that the previously identified conidiation genes dewA, fluG, and stuA may function in isotropic expansion during early vegetative growth and asexual reproduction.
Except for a brief period of isotropic expansion after spores break dormancy, filamentous fungi grow exclusively by adding new material to the tips of long tubular cells (hyphae). This highly polar tip extension allows filamentous fungi to penetrate substrates and is essential to their roles as saprophytes breaking down organic material in the environment and as pathogens invading plant and animal tissues. Some of the genes required for polar growth have been identified in the model filamentous fungus Aspergillus nidulans by analyzing temperature-sensitive mutants defective in germ tube emergence at 42°C (4, 11, 14) and by reverse genetics (5).
In A. nidulans, following the breaking of dormancy, the asexual spore (conidium) expands isotropically, with cell wall components added uniformly in all directions until after the first nuclear division (5 to 6 h under typical conditions), when a switch to polar growth occurs that results in the emergence of a nascent germ tube (10). The germ tube continues to elongate and branches, eventually forming a hyphal network (mycelium). After 24 h, under typical conditions, asexual reproduction is initiated and aerial conidiophore stalks extend from specialized foot cells of the mycelium. These give rise to differentiated multicellular conidiophores, which produce long chains of uninucleate conidia, the final product of asexual reproduction. Here we use microarray analysis to identify transcriptional changes in the early vegetative growth of A. nidulans.
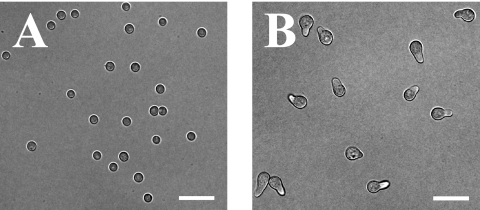
Flasks of 300 ml complete medium plus uracil and uridine (CMUU) (11) were inoculated with 10 ml wild-type (strain A773) spore stock (108 spores/ml) and incubated for 3 or 5 h at 42°C with constant shaking (250 rpm). The temperature of 42°C was chosen to facilitate later comparison with temperature-sensitive mutants. Microscopic inspection after 3 h of incubation revealed that 100% of spores lacked germ tubes, while after 5 h of incubation, 90% had early germ tubes (n = 200) (Fig. 1). Following incubation, cells were harvested using a Steritop filter (Millipore), frozen, ground in liquid nitrogen, and RNA purified using TRIzol according to the manufacturer's instructions (Invitrogen). cDNA synthesis and indirect Cy dye incorporation were performed using the standard TIGR protocol (standard operating procedure [SOP] M007; http://pfgrc.tigr.org/protocols/protocols.shtml) with the following modifications. Five micrograms of total RNA was used in the first-strand cDNA reaction with a 3:1 aminoallyl-dUTP-to-dTTP ratio. Dried aminoallyl-labeled cDNA was resuspended in 9 μl of 0.05 M sodium carbonate (pH 9.0) and added directly to the appropriate dye vial from the Amersham CyDye postlabeling reactive dye pack (RPN5661). The resulting labeled cDNA was hybridized to an A. nidulans glass slide microarray spotted with 23,962 70-mers representing all predicted open reading frames (PFGRC/TIGR A. nidulans microarray, version 1; a full description and annotation are available at Gene Expression Omnibus [accession no. GPL5138; http://www.ncbi.nlm.nih.gov/geo/]) according to the standard TIGR protocol (SOP M008). Two biological replicates were performed using cultures grown in parallel, and technical “dye swap” replicates were carried out for each biological replicate for a total of four hybridizations. The slides were scanned using a Perkin-Elmer ScanArray microarray scanner, and the resulting tagged-image format file images were imported into TIGR Spotfinder (version 3.1.1). Raw intensities were calculated for each detectable spot by using the Otsu method, and quality control filtering was used to eliminate values from spots with poor morphology or low signal-to-noise ratios. The resulting intensities were then imported into TIGR MIDAS (version 2.20), and those with values of <10,000 were removed from the data set by using the low-intensity filter. Normalization was done using the LOWESS (Locfit) algorithm (global mode; 0.33 smoothing parameter), and print tip bias was addressed by using standard deviation regularization. The dye swap consistency filter was used to remove data for genes with inconsistent expression levels between dye swap replicates. Finally, the in-slide replicate function was used to average the normalized intensity values representing replicate spots on the array. The resulting data were analyzed using Microsoft Excel, and genes that had at least a twofold difference in expression were scored as differentially regulated. The entire set of supporting microarray data is available at Gene Expression Omnibus (accession no. GSE7698).
FIG. 1.
Microarray analysis was used to compare isotropically expanding cells (3 h of incubation) (A) and polarly extending cells (5 h of incubation) (B). Scale bars, 20 μm.
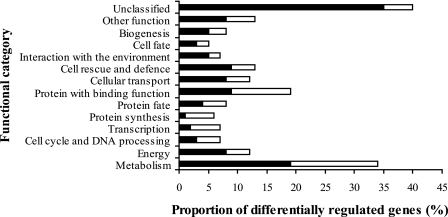
A total of 315 genes were differentially regulated following the switch from isotropic to polar growth (218 were up-regulated and 97 were down-regulated). Differentially expressed genes were classified into functional groups (Fig. 2) by using their assigned MIPS functional catalog (FunCat) annotation in the PEDANT A. nidulans genome database (http://pedant.gsf.de/). FunCat currently contains 27 main categories that focus on functional processes (12). The categorized genes represent a diverse range of functions, and the most prominent group is metabolism related. Interestingly, 40% of the genes that are differentially regulated during the isotropic-to-polar switch are unclassified and therefore represent candidates for novel polarity-related genes.
FIG. 2.
Functional classification of genes that are differentially regulated during germ tube formation in A. nidulans. Differentially regulated genes were classified into functional groups using the PEDANT genome database and the MIPS FunCat. Many genes were assigned to multiple functional categories. Solid bars represent genes that were up-regulated more than twofold, while open bars represent those down-regulated more than twofold.
The 10 genes with the greatest up-regulation and the 10 with the greatest down-regulation during early vegetative growth are shown in Table 1. Surprisingly, transcripts representing the conidiation genes dewA, fluG, and stuA (1, 8, 16) were detected during the vegetative isotropic-to-polar growth switch; dewA and fluG were the second and fourth most highly down-regulated genes (5.0-fold and 4.3-fold, respectively), and stuA showed no significant change in its transcript level. Transcripts of the known conidiation genes brlA, abaA, wetA, and rodA were not detected in either the isotropic or the polar growth stage. dewA encodes a fungal hydrophobin component of the spore wall and has previously been shown to be transcribed specifically during conidiation (16). fluG encodes a protein that appears to be involved in the generation of an extracellular signal responsible for the activation of asexual reproduction and has previously been shown to be constitutively expressed during late vegetative growth and asexual reproduction (6, 13). stuA encodes a helix-loop-helix transcription factor required for proper differentiation and organization of the conidiophore and has previously been shown to be up-regulated during late vegetative growth, with further up-regulation during asexual reproduction (8, 9, 17).
TABLE 1.
The top 10 up-regulated and the top 10 down-regulated genes during early vegetative growth in A. nidulans
| Locus | A. nidulans annotation or best informative BLAST match (accession no.; E value)a | Fold change in expressionb |
|---|---|---|
| Up-regulated genes | ||
| AN8638.3 | Neosartorya fischeri hemerythrin HHE cation binding domain protein (XP_001261781; 1e−67) | +17.0 |
| AN11337.3 | No informative BLAST hits | +13.8 |
| AN8124.3 | Cryptococcus neoformans var. neoformans membrane protein (XP_572852; 7e−78) | +12.9 |
| AN6274.3 | Neosartorya fischeri short-chain dehydrogenase (XP_001264016; 9e−125) | +11.8 |
| AN6693.3 | No informative BLAST hits | +9.8 |
| AN2683.3 | Aspergillus clavatus integral membrane protein (XP_001274701; 6e−99) | +9.5 |
| AN8647.3 | Neosartorya fischeri formate/nitrite transporter, putative (XP_001262468; 8e−121) | +9.3 |
| AN1659.3 | Aspergillus terreus amino acid permease INDA1 (XP_001214474; 0.0) | +8.6 |
| AN11574.3 | No informative BLAST hits | +8.3 |
| AN0726.3 | Neosartorya fischeri SUN domain protein Adg3, putative (XP_001264362; 1e−136) | +8.0 |
| Down-regulated genes | ||
| AN3032.3 | Aspergillus clavatus cell surface protein, putative (XP_001270917; 9e−14) | −5.9 |
| AN4819.3 | A. nidulans FluG | −5.0 |
| AN6794.3 | No informative BLAST hits | −4.8 |
| AN8006.3 | A. nidulans DewA hydrophobin | −4.3 |
| AN6368.3 | Aspergillus fumigatus arginyl-tRNA synthetase (XP_755736; 0.0) | −4.2 |
| AN7540.3 | Aspergillus fumigatus eukaryotic translation initiation factor 3 subunit EifCd (XP_755799; 0.0) | −3.4 |
| AN5155.3 | Neosartorya fischeri tRNA-specific adenosine-34 deaminase subunit Tad3, putative (XP_001264959; 9e−122) | −3.2 |
| AN3800.3 | No informative BLAST hits | −3.1 |
| AN4770.3 | Phosphoadenosine phosphosulfate reductase | −3.1 |
| AN8269.3 | Aspergillus terreus heat shock protein 82 (XP_001216617; 0.0) | −3.0 |
BLASTX searches were used for genes designated “hypothetical” in the A. nidulans genome database (Broad Institute). Genes with E values of >1e−5 are designated “No informative BLAST hits.”
Derived from the mean of two biological replicates.
Though many previous studies have characterized the expression of conidiation genes during the transition from vegetative polar growth to asexual reproduction, none have characterized expression in early vegetative growth. To determine whether the expression of conidiation genes in our microarray experiments was consistent with the findings of previously published studies, we compared gene expression before and after the induction of conidiation. Synchronous asexual development is induced in A. nidulans cells grown in submerged culture by exposing them to an air interface (9). Flasks of 300 ml CMUU were inoculated with 108 conidia and grown with continual shaking (250 rpm) for 18 h at 42°C. The resulting undifferentiated hyphae were either harvested or transferred to filters and incubated on a solid medium for 24 h at 42°C to induce synchronous conidiation. RNA was harvested from both undifferentiated hyphae and synchronously conidiating cultures and was used in microarray analysis as described above. The conidiation genes brlA, abaA, wetA, dewA, rodA, and stuA were all up-regulated in conidiating cultures, in agreement with previously published work (3, 9, 15, 16). fluG was expressed both before and after the induction of conidiation with no significant change in its mRNA level, a finding also consistent with previous studies (1).
Transcripts of genes expressed at high levels in conidiating cultures might be packaged into conidia and survive for long periods of time. To determine whether dewA, fluG, and stuA mRNAs detected during early vegetative isotropic growth represent RNAs packaged into the conidium or new transcription, RNA was extracted from isolated dormant conidia, labeled, and hybridized with RNA isolated from isotropically expanding cells as described above. The dewA transcript level was 4.5-fold higher in dormant conidia than in isotropically expanding cells, raising the possibility that dewA mRNA detected in isotropically expanding cells might represent RNA that was packaged into the conidium and survived dormancy. In contrast, fluG and stuA transcripts were not detected in dormant conidia, demonstrating that these conidiation genes are transcribed early in vegetative growth.
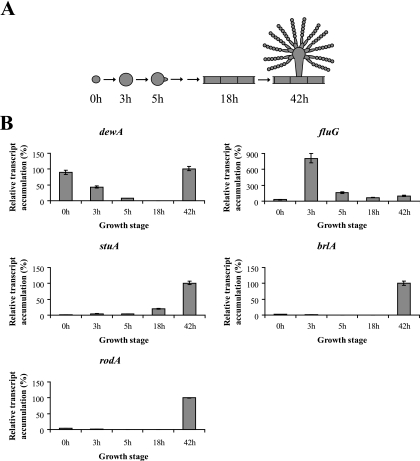
To determine the relative levels of transcripts and to validate the microarray data, we used quantitative real-time PCR (RT-PCR) to measure the transcript levels of the conidiation genes dewA, fluG, stuA, brlA, and rodA (Fig. 3). All RT-PCRs were carried out using an Applied Biosystems 7500 system with SYBR green detection. The thermocycling conditions consisted of 2 initial incubations of 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Three technical replicates were carried out for each reaction, and the data were normalized according the 2−ΔΔCT method (7) using 18S rRNA as the internal reference. The RT-PCR results for all genes were consistent with the microarray data. Remarkably, the transcript level for fluG during isotropic growth was fivefold higher than that at any other time examined (Fig. 3). In contrast, the transcript level for stuA during isotropic growth was 4-fold greater than that in dormant conidia but 20-fold lower than that in conidiating cultures.
FIG. 3.
Quantitative RT-PCR of A. nidulans conidiation genes during early vegetative growth and asexual reproduction. (A) Time points represent ungerminated conidia (0 h), isotropically expanding cells (3 h), polarly extending cells (5 h), mature vegetative hyphae (18 h), and synchronously conidiating cultures with mature conidia (42 h). (B) Transcript accumulation for each gene was plotted relative to the level at 42 h (taken as 100%). The data were normalized according to the 2−ΔΔCT method using 18S rRNA as the internal reference. Error bars represent ranges of three technical replicates.
Our data show that transcripts for dewA, fluG, and stuA, three genes previously shown to be important for conidiation, are present during early vegetative growth. Our data also show that two of these genes, fluG and stuA, are actively transcribed during isotropic growth and that dewA and fluG are differentially regulated during the isotropic-to-polar growth switch. Further work is required to establish if these conidiation genes have significant biological functions in isotropic expansion during early vegetative growth. The hydrophobin encoded by dewA has previously been shown to be specifically transcribed during the late stages of conidiation, when nascent asexual spores bud from the conidiophore, a process involving isotropic expansion (16). stuA has also previously been shown to be up-regulated during asexual reproduction. A fivefold increase in the transcript level coincided with conidiophore vesicle formation (9), another growth stage involving isotropic expansion. FluG is required for the activation of conidiation and the production of the mycotoxin sterigmatocystin (6, 13), a secondary metabolite whose production is coordinated with asexual sporulation (2). Although fluG deletion mutants produce aerial hyphae, they fail to differentiate proper conidiophore stalks with swollen vesicles. Our results suggest that dewA, fluG, and stuA may function in isotropic expansion during both vegetative growth and asexual reproduction.
Acknowledgments
This work was supported by DOE Biosciences grant DE-FG02-97ER20275 to M.M.
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Adams, T. H., W. A. Hide, L. N. Yager, and B. N. Lee. 1992. Isolation of a gene required for programmed initiation of development by Aspergillus nidulans. Mol. Cell. Biol. 12:3827-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, T. H., and J. H. Yu. 1998. Coordinate control of secondary metabolite production and asexual sporulation in Aspergillus nidulans. Curr. Opin. Microbiol. 1:674-677. [DOI] [PubMed] [Google Scholar]
- 3.Boylan, M. T., P. M. Mirabito, C. E. Willett, C. R. Zimmerman, and W. E. Timberlake. 1987. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol. Cell. Biol. 7:3113-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris, S. D., A. F. Hofmann, H. W. Tedford, and M. P. Lee. 1999. Identification and characterization of genes required for hyphal morphogenesis in the filamentous fungus Aspergillus nidulans. Genetics 151:1015-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris, S. D., and M. Momany. 2004. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41:391-400. [DOI] [PubMed] [Google Scholar]
- 6.Lee, B. N., and T. H. Adams. 1994. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 8:641-651. [DOI] [PubMed] [Google Scholar]
- 7.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 8.Miller, K. Y., T. M. Toennis, T. H. Adams, and B. L. Miller. 1991. Isolation and transcriptional characterization of a morphological modifier: the Aspergillus nidulans stunted (stuA) gene. Mol. Gen. Genet. 227:285-292. [DOI] [PubMed] [Google Scholar]
- 9.Miller, K. Y., J. Wu, and B. L. Miller. 1992. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 6:1770-1782. [DOI] [PubMed] [Google Scholar]
- 10.Momany, M., and I. Taylor. 2000. Landmarks in the early duplication cycles of Aspergillus fumigatus and Aspergillus nidulans: polarity, germ tube emergence and septation. Microbiology 146:3279-3284. [DOI] [PubMed] [Google Scholar]
- 11.Momany, M., P. J. Westfall, and G. Abramowsky. 1999. Aspergillus nidulans swo mutants show defects in polarity establishment, polarity maintenance and hyphal morphogenesis. Genetics 151:557-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruepp, A., A. Zollner, D. Maier, K. Albermann, J. Hani, M. Mokrejs, I. Tetko, U. Guldener, G. Mannhaupt, M. Munsterkotter, and H. W. Mewes. 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32:5539-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seo, J. A., Y. Guan, and J. H. Yu. 2006. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 172:1535-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi, X., Y. Sha, and S. Kaminskyj. 2004. Aspergillus nidulans hypA regulates morphogenesis through the secretion pathway. Fungal Genet. Biol. 41:75-88. [DOI] [PubMed] [Google Scholar]
- 15.Stringer, M. A., R. A. Dean, T. C. Sewall, and W. E. Timberlake. 1991. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 5:1161-1171. [DOI] [PubMed] [Google Scholar]
- 16.Stringer, M. A., and W. E. Timberlake. 1995. dewA encodes a fungal hydrophobin component of the Aspergillus spore wall. Mol. Microbiol. 16:33-44. [DOI] [PubMed] [Google Scholar]
- 17.Wu, J., and B. L. Miller. 1997. Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol. Cell. Biol. 17:6191-6201. [DOI] [PMC free article] [PubMed] [Google Scholar]