Abstract
The parasitic protozoan Entamoeba histolytica relies on a very dynamic cytoskeleton in order to invade and survive in host tissues. Identification of cytoskeletal elements is key to understanding these processes. Here we present the characterization of EhLimA, the first LIM protein of E. histolytica. EhLimA consists of a single LIM domain at its N terminus and exhibits the highest degree of homology with DdLimE from Dictyostelium discoideum. Immunofluorescence localization of EhLimA using anti-EhLimA antibodies revealed that EhLimA is highly concentrated at the plasma membrane of cells. Silencing or overexpression of the EhLimA gene did not have a significant effect on the growth or morphology of the parasite. EhLimA associates with the cytoskeleton as demonstrated by the enrichment of the protein in cytoskeleton fractions as well as in pull-down assays that revealed that cytoskeleton association involves interaction with actin. EhLimA binding to actin was shown to be dependent on the N-terminal LIM domain of EhLimA, as removal of even half of the LIM domain resulted in almost complete inhibition of the binding to actin. We also found that a portion of EhLimA floats to the lower-density regions of a sucrose gradient together with portions of the Gal-lectin light subunit and actin. Treatment of cells with the cholesterol-sequestering agent digitonin resulted in increased solubility of EhLimA. These results indicate that in addition to cytoskeletal association, EhLimA may also associate with lipid rafts in the parasite plasma membrane and suggest that EhLimA may be part of the molecular system connecting the actin cytoskeleton to membrane rafts.
Many cellular functions are dependent on specific protein-protein interactions. These protein-protein interactions are governed by a variety of protein binding domains one of which is the LIM domain. Since first described nearly 2 decades ago (21, 28, 57), LIM domain-containing proteins have been identified in a wide range of eukaryotes, including protozoa (16, 30, 45), plants (5, 20, 40), and yeast (Saccharomyces cerevisiae) (39), stressing the evolutionary importance of these proteins. In the parasitic protozoan Entamoeba histolytica, a few studies have revealed the presence of such proteins (17, 36), although to date, no LIM domain-containing proteins have been characterized in this organism. The name LIM is derived from the three developmentally regulated transcription factors, LIN-11 of Caenorhabditis elegans (21), Isl1 of rat (28), and MEC-3 of C. elegans (57) in which the LIM domain was initially identified. The LIM domain is a cysteine-rich motif consisting of two zinc finger-like modules and displaying the consensus sequence CX2CX16-23HX2CX2CX2CX16-21CX2(C/H/D/) (31). This domain is found in a variety of different proteins with diverse functions, including transcription factors and cytoskeleton-associated proteins, and may be found in association with other functional domains, such as homeodomains, protein kinase domains, or other protein binding domains (31, 54). It serves as a protein binding interface capable of associating with a wide range of proteins and mediating protein-protein interactions. The various binding partners of LIM domains influence the subcellular localization and functions of the LIM proteins. It is therefore not surprising that LIM domain-containing proteins have been found to participate in a broad range of biological processes, including cytoskeleton organization, transcriptional regulation, development of cell types, and signaling (4, 18, 31).
LIM domain-containing proteins do not form a functional family but have been classified into three groups based on the sequence relationships among the LIM domains and overall structure of the proteins. Group 1 proteins are found primarily in the nucleus and include LIM homeodomain proteins and LIM-only proteins that are involved in transcriptional regulation. Group 2 proteins are composed primarily of LIM domains and include members of the cysteine-rich protein family. Group 3 proteins contain different numbers of LIM domains located at the C terminus and include proteins such as zyxin and paxillin. Group 2 and group 3 proteins are primarily cytoplasmic, and many are associated with the actin cytoskeleton (18).
In Dictyostelium discoideum, a number of LIM domain-containing proteins have been characterized (16, 30, 45). The known similarity between several proteins of this amoeba with those of E. histolytica prompted us to investigate whether analogous LIM domain-containing proteins exist in E. histolytica. Here we present the characterization of the first LIM domain-containing protein in E. histolytica which we termed E. histolytica LimA (EhLimA) (NCBI accession XP_656918.1). We show that EhLimA associates with the actin cytoskeleton and possibly with lipid raft domains in the plasma membrane, suggesting that it may serve to connect the actin cytoskeleton to membrane rafts.
MATERIALS AND METHODS
Strains and culture conditions.
Trophozoites of E. histolytica strain HM-1:IMSS were grown at 37°C in TYI-S-33 medium (19). Transfection of trophozoites was performed as previously described (24). Transfectants were grown in the presence of the neomycin derivative G418.
Cloning of EhLimA cDNA and sequence analysis.
The EhLimA gene sequence was obtained from the E. histolytica Genome Project (35) of The Institute for Genomic Research (TIGR) and The Wellcome Trust Sanger Institute. The gene was cloned into the pBluescript II KS vector following PCR amplification using cDNA as the template and primers A and B (Table 1). These primers were also used to sequence the gene in both orientations. The sequence obtained was in complete accordance with the sequence of the gene appearing in the E. histolytica Genome Project (35).
TABLE 1.
Primers used for construction of plasmids
| Primer | Primer sequence | Orientationa |
|---|---|---|
| A | 5′-ACGGATCCAATGTCTGCTAAGAAGTGTT | S |
| B | 5′-TTAAGTCGACTTAAAAACCTTCTTCTTCTTC | AS |
| C | 5′-AAGGTCTCCCATGTCTGCTAAGAAGTGTTTTGCT | S |
| D | 5′-CATGCCATGGATTATAAAGATGATGATGATAAATCTGCTAAGAAGTGTTTTGCT | S |
| E | 5′-CATGCCATGGATTATAAAGATGATGATGATAAATTCAAATGTAAAGAATGTGGTC | S |
| F | 5′-GCGTGTCGACTTAATGTGGTTTTCCTCCAGCTC | AS |
| G | 5′-CATGCCATGGCTTCTGCTAAGAAGTGTTTTG | S |
| H | 5′-GCGTGTCGACTTATTTATCATCATCATCTTTATAATCAAAACCTTCTTCTTCTTCATG | AS |
| I | 5′-CATGCCATGGCGTCTGCTAAGAAGTGTTTTG | S |
| J | 5′-TCCCCGCGGCTTGCTGCACCCTTTG | S |
| K | 5′-CATGCCATGGTCATGATTGTTTGTAAGATATG | AS |
| L | 5′-CGGGGTACCTATGTCTGCTAAGAAGTGTTTTG | S |
| M | 5′-CGCGGATCCTTAAAAACCTTCTTCTTCTTC | AS |
| N | 5′-GACTCGAGTCGACATCGATT(T16) | AS |
S, sense; AS, antisense.
Sequence analysis of EhLimA was performed using various tools available on the ExPASy server (www.expasy.ch/tools/). Multiple alignment was performed using the ClustalW multiple-sequence alignment program (www.ebi.ac.uk/clustalw/).
Construction of plasmids for overexpression.
All plasmid constructs intended for overexpression were generated using an intermediate plasmid as follows. The sequence of interest was first cloned into the NcoI and SalI restriction sites of the pSA21 intermediate plasmid construct (29). The pSA21 plasmid construct containing the sequence of interest was then digested with BamHI and SacI, which releases a cassette containing the sequence of interest flanked by 5′ and 3′ actin regulatory sequences. This cassette was then transferred to the pEhAct-Neo (38) vector, after removal of its cassette by digestion with BamHI and SacI.
A plasmid containing the full-length EhLimA sequence (438 bp) was first prepared by PCR amplification using E. histolytica cDNA as the template and primers C and B (Table 1). This plasmid was then used as the template for preparing the sequence of interest as described below. N-terminally and C-terminally FLAG-tagged EhLimA sequences were obtained by PCR amplification using the plasmid containing the full-length EhLimA sequence (described above) as the template. The 24 nucleotides corresponding to the FLAG epitope (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys) were synthesized as part of either the 5′ or 3′ primer for N-terminally or C-terminally tagged EhLimA, respectively, and in such a way that they were introduced into the sequence. For full-length EhLimA tagged at the N terminus (462 bp), primers D and B (Table 1) were used. For N-terminus-truncated EhLimA tagged at the N terminus (375 bp), primers E and B (Table 1) were used. For C-terminus-truncated EhLimA tagged at the N terminus (327 bp), primers D and F (Table 1) were used. For full-length EhLimA tagged at the C terminus (465 bp), primers G and H (Table 1) were used.
Construction of a plasmid for transcriptional silencing.
Transcriptional silencing of EhLimA was achieved as previously described for other genes (7) in the G3 strain of E. histolytica that is already silenced in the expression of the E. histolytica amoebapore gene (Ehap-a) (8). The full-length EhLimA sequence was obtained by PCR amplification using the plasmid containing the full-length EhLimA (described above) as the template and primers I and B (Table 1) containing NcoI and SalI sites, respectively. The 5′ upstream segment (473 bp) of the Ehap-a gene was obtained by PCR amplification using plasmid psAP-1 (7) as the template and primers J and K (Table 1) containing SacII and NcoI sites, respectively. Both of these PCR products were digested with NcoI and ligated to each other with T4 DNA ligase. The ligation product was amplified by PCR using primers J and B (Table 1). The PCR product obtained was digested with SacII and SalI and ligated to the 3′ Ehactin flanking region that was obtained by digestion of the pSA21 (29) plasmid with SalI and BamHI. This cassette containing the 5′ upstream segment of the Ehap-a gene, the full-length EhLimA sequence, and the 3′ Ehactin flanking region was cloned into the pEhAct-Neo (38) vector digested at SacII and BamHI. The resulting plasmid was transfected into the G3 strain, and transfectants were selected as previously described (7).
Reverse transcription-PCR (RT-PCR).
Total RNA was prepared using the TRI reagent RNA isolation kit (Sigma). Five micrograms of total RNA was reverse transcribed using an oligo(dT) adaptor primer (Table 1, primer N) and avian myeloblastosis virus reverse transcriptase (Promega) according to the manufacturer's protocol. The cDNA product was diluted 25 times and used as the template for subsequent PCRs.
Production and purification of polyclonal antibodies.
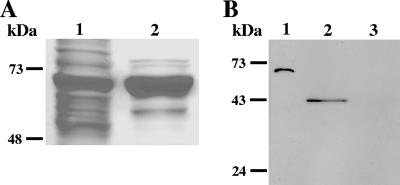
In order to produce specific antibodies against EhLimA, the gene was cloned into a pET bacterial expression vector downstream of the highly expressed bacterial xylanase protein fused to an amino-terminal hexahistidine tag (a kind gift from Yoav Barak). EhLimA was amplified by PCR using E. histolytica cDNA as the template and primers L and M (Table 1). The PCR product obtained was digested with KpnI and BamHI and cloned into the corresponding sites of the pET vector. The His-tagged xylanase-EhLimA fusion protein was expressed in Escherichia coli (Fig. 1A) and purified by affinity chromatography using Ni-nitrilotriacetic acid agarose (QIAGEN). The purified fusion protein was separated on a polyacrylamide gel, and the band representing xylanase-EhLimA was sliced from the gel and fragmented by passing it repeatedly through a syringe. Two rabbits were injected five times with 100 μg of protein per injection and an interval of 3 weeks between each injection. For the first injection, the gel was emulsified with complete Freund's adjuvant, and for the second injection, the gel was emulsified with incomplete Freund's adjuvant. For the remaining injections, phosphate-buffered saline (PBS) was used. Purified anti-EhLimA antibodies were obtained by ammonium sulfate precipitation followed by passing the antibody mixture through a column of Sepharose beads (a kind gift from Talia Miron) coupled to the purified xylanase protein (a kind gift from Yoav Barak). The antixylanase antibodies bound to the column, while the anti-EhLimA antibodies were eluted. The specificity of the anti-EhLimA antibodies is shown in Fig. 1B. Purified anti-EhLimA antibodies were divided into aliquots and stored at −20°C until being utilized for Western blotting and immunofluorescence labeling.
FIG. 1.
(A) Analysis of the His-tagged xylanase-EhLimA by gel electrophoresis following expression and purification in E. coli. Lane 1, total lysate of bacterial cells overexpressing the His-tagged xylanase-EhLimA fusion protein; lane 2, purified xylanase-EhLimA fusion protein. (B) A Western blot analysis demonstrating the specificity of the purified anti-EhLimA antibodies. The purified xylanase-EhLimA fusion protein (lane 1) and a purified glutathione S-transferase (GST)-EhLimA fusion protein (lane 2) are recognized by the antibodies, while purified GST (lane 3) is not.
Preparation of protein fractions.
Total cell lysates of E. histolytica trophozoites were obtained by suspending PBS-washed trophozoites in lysis buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and protease inhibitors 0.2 mM leupeptin, 5 mM iodoacetamide, 5 mM 1,10-phenanthroline monohydrate, and 2 mM phenylmethylsulfonyl fluoride) and incubating for 30 min on ice. The lysates were subjected to centrifugation at 13,000 × g in a microcentrifuge at 4°C, and after removal of the supernatant, the pellet was resuspended in lysis buffer. The protein concentration of each fraction was determined by the Bradford assay (9), and 5 to 10 μg of protein was taken for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blots.
SDS-PAGE and Western blots.
Samples were prepared by the addition of sample buffer and boiling for 3 min. Samples were separated on a 15% polyacrylamide gel under reducing conditions. Gels were either stained for total protein with GelCode blue stain reagent (Pierce) or blotted onto nitrocellulose membranes. Membranes were stained for total protein with Ponceau S before incubation with antibodies. The dilutions of primary antibodies used in this study were as follows: polyclonal anti-EhLimA, 1:500; monoclonal anti-FLAG, 1:2,000 (Sigma); monoclonal antiactin, 1:1,000 (ICN Biomedicals, Inc.); and polyclonal anti-Gal-lectin light subunit, 1:1,000 (3). These were followed by incubations with either a horseradish peroxidase-conjugated goat anti-rabbit antibody (1:10,000; Jackson) or a horseradish peroxidase-conjugated goat anti-mouse antibody (1:5,000; Jackson). Blots were developed with an enhanced chemiluminescence kit (ECL; Amersham).
Immunofluorescence and confocal microscopy.
E. histolytica cells were resuspended in warm TYI-S-33 medium, set on coverslips in a petri dish, and left to adhere for 10 min at 37°C. After removal of the medium, cells were fixed with prewarmed PBS containing 3.7% formaldehyde (3.7% formaldehyde-PBS) for 30 min. The coverslips were washed with PBS and permeabilized with PBS containing 0.1% Triton X-100 for 3 min. Coverslips were washed with PBS and blocked with PBS containing 5% BSA (5% BSA-PBS) for 1 h. Purified polyclonal anti-EhLimA antibodies were added at a dilution of 1:10 in 5% BSA-PBS and incubated for 1 h. The coverslips were washed with PBS and incubated for 30 min with fluorescein isothiocyanate-conjugated goat anti-rabbit antibodies (Sigma) at a dilution of 1:200 in 5% BSA-PBS. For actin staining, cells were grown overnight on coverslips and fixed with prewarmed 3.7% paraformaldehyde-PBS. Blocking was performed for 30 min with 1% BSA-PBS followed by 40 min of incubation with phalloidin-tetramethyl rhodamine isocyanate (TRITC) (1 mg/ml; Sigma) at a dilution of 1:200 in 1% BSA-PBS. Coverslips were washed in PBS and mounted on a glass slide using VECTASHIELD mounting medium (VECTOR Laboratories). The coverslips were sealed in place with nail polish. Fluorescent samples were viewed on a confocal laser-scanning microscope using either a 40× or 60× objective.
Preparation of cytoskeletal fractions.
E. histolytica trophozoites were harvested, washed, resuspended in lysis buffer (60 mM PIPES, 25 mM HEPES, 125 mM KCl, 2 mM MgCl2, 5 mM EGTA, 0.5 mM ATP, pH 7.2, 1% Triton X-100, and protease inhibitors 0.2 mM leupeptin, 5 mM iodoacetamide, 5 mM 1,10-phenanthroline monohydrate, and 2 mM phenylmethylsulfonyl fluoride) and incubated for 30 min on ice. Lysates were centrifuged at 4°C for 15 min at 500 × g. The supernatant was subjected to ultracentrifugation at 4°C for 1 h at 100,000 × g. The supernatant, which contained the cytoplasmic proteins as well as membrane proteins that were solubilized, was transferred to a new tube. The pellet, which contained the cytoskeleton and associated proteins, as well as other Triton X-100-insoluble proteins, such as those associated with lipid rafts, was resuspended in the above buffer. The protein concentration was determined by the Bradford assay (9).
Immunoprecipitation of FLAG-tagged fusion proteins and immunodetection of actin.
E. histolytica cells overexpressing the various FLAG-tagged EhLimA fusion proteins as well as control cells were lysed, and lysates were centrifuged at 4°C for 10 min at 13,000 × g. The supernatants were incubated with anti-FLAG M2 affinity gel (Sigma) on a roller shaker at 4°C for 2 h. Resin-bound FLAG-tagged fusion proteins were pelleted, and FLAG-tagged proteins were eluted by the addition of 0.1 M glycine HCl. Purified FLAG-tagged fusion proteins were visualized following SDS-PAGE and total protein staining as described above. SDS-PAGE, followed by immunoblotting with antiactin antibodies as described above, was performed in order to detect coimmunoprecipitation of actin.
Treatment of cells with methyl-β-cyclodextrin or digitonin.
For cholesterol depletion, amoeba cells were incubated with either 20 mM methyl-β-cyclodextrin or with 0.2% digitonin in PBS for 30 min on ice. Following incubation, cells were lysed with lysis buffer containing Triton X-100, and soluble and insoluble fractions were obtained as described above.
Sucrose gradient flotation.
E. histolytica trophozoites were lysed in 1 ml of cold morpholineethanesulfonic acid (MES)-buffered saline (25 mM MES, pH 6.5, 150 mM NaCl, 1% Triton X-100, and protease inhibitors 0.2 mM leupeptin, 5 mM iodoacetamide, 5 mM 1,10-phenanthroline monohydrate, 2 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, and 1 mM sodium pyrophosphate) and incubated for 30 min on ice. Total cell lysates were adjusted to 40% sucrose by adding an equal volume of 80% sucrose and placed at the bottom of an ultracentrifuge tube. A discontinuous sucrose gradient was generated by overlaying this with 30% sucrose and 5% sucrose. Tubes were centrifuged at 4°C for 20 h at 260,000 × g (39,000 rpm) in an SW41 rotor. Following centrifugation, 12 fractions of 1 ml each were collected from the top of the gradient. Proteins were precipitated by the addition of trichloroacetic acid. Protein pellets were resuspended in sample buffer and subjected to SDS-PAGE and immunoblotting as described above. The percentage of sucrose in each fraction was determined using a refractometer (Bausch & Lomb).
RESULTS
Cloning and sequence analysis of a LIM domain-containing protein in E. histolytica.
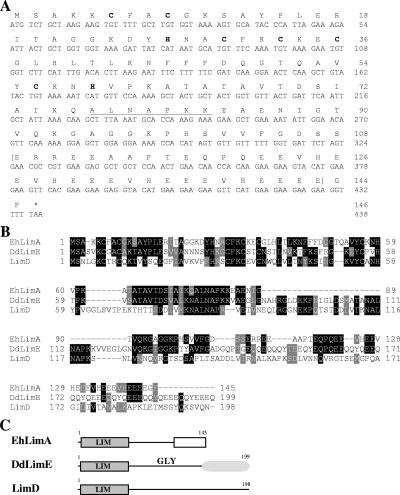
The gene encoding a LIM domain-containing protein in E. histolytica was cloned, and the corresponding protein was termed EhLimA (NCBI accession no. XP_656918.1). The sequence of the EhLimA gene is 438 nucleotides in length and codes for a protein of 145 amino acids with a calculated molecular mass of 16 kDa. Sequencing data obtained were in complete accordance with the sequence of the gene that appears in the E. histolytica genome (35). Analysis of the deduced amino acid sequence of EhLimA revealed a single LIM domain at its N terminus spanning amino acid residues 6 to 59 (Fig. 2A). The C terminus of the EhLimA protein is rich in glutamic acid, and 18 of the 22 glutamic acid residues are found in the C terminus. EhLimA exhibits the highest degree of homology (37% identity and 47% similarity) to DdLimE (Fig. 2B and C), a Dictyostelium discoideum LIM domain protein that also has a single LIM domain at its N terminus (45, 48). The LIM domains of EhLimA and DdLimE are 44% identical. The glycine-rich domain that follows the LIM domain and the C-terminal coiled-coil region found in DdLimE are not present in the EhLimA sequence. However, EhLimA contains part of a repeat motif, ALNAPKK, that appears twice in DdLimE. EhLimA was also found to be somewhat homologous to Dictyostelium LimD (19% identity and 28% similarity), that also consists of a single LIM domain at its N terminus (30). Both DdLimE and LimD of Dictyostelium have been classified with group 2 of LIM domain-containing proteins. Similarly, we propose that EhLimA be classified with this group of proteins as well.
FIG. 2.
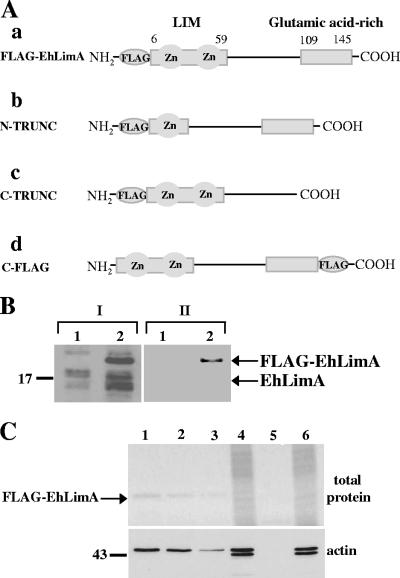
(A) Nucleotide sequence and deduced amino acid sequence of EhLimA. The conserved cysteine and histidine residues of the N-terminal LIM domain are shown (bold letters). The glutamic acid-rich C terminus of the protein is indicated by brackets. Part of the repeat motif that appears twice in Dictyostelium DdLimE is underlined. (B) Alignment of the EhLimA amino acid sequence with the amino acid sequences of Dictyostelium DdLimE and LimD. Identical residues are represented by black boxes, and similar residues are represented by gray boxes. Sequences were aligned using the ClustalW multiple-sequence alignment program (www.ebi.ac.uk/clustalw/). GenBank accession numbers are as follows: EhLimA, XP_656918; DdLimE, U97699; LimD, AF348467. (C) Schematic representation of EhLimA and Dictyostelium proteins DdLimE and LimD. The single N-terminal LIM domain present in all proteins is indicated. The glutamic acid-rich region of EhLimA is represented by a white box. The glycine-rich region (GLY) and coiled-coil region (shaded oval) of DdLimE are shown.
Expression of EhLimA in E. histolytica.
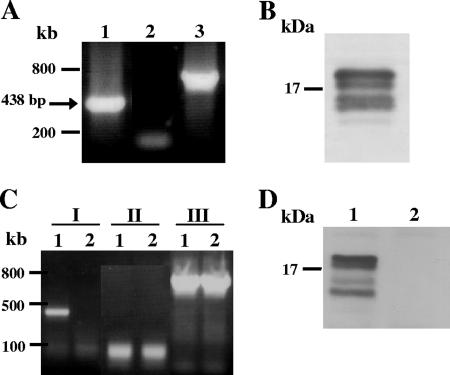
Transcription of EhLimA was verified in an RT-PCR analysis of total RNA. Amplification of a band of the expected size of 438 bp (Fig. 3A) was obtained, indicating that EhLimA is transcribed in E. histolytica. Protein expression was analyzed in a Western blot using purified polyclonal antibodies prepared against recombinant EhLimA (Fig. 3B). The anti-EhLimA antibodies specifically recognize four bands around 16 kDa in size in amoeba total cell lysates.
FIG. 3.
mRNA and protein expression of EhLimA. (A) An RT-PCR analysis of total RNA using primers specific for EhLimA results in amplification of a band of the expected size of 438 bp (lane 1). No amplification is attained when the reverse transcription reaction is performed without reverse transcriptase (lane 2). Amplification of actin is shown in lane 3. (B) Detection of EhLimA in a Western blot analysis of total cell lysates using purified anti-EhLimA antibodies reveals four bands around 16 kDa. (C) An RT-PCR analysis of total RNA of G3 transfected trophozoites in which EhLimA is transcriptionally silenced (lane 2) and of nontransfected G3 cells expressing EhLimA (lane 1). Amplification of EhLimA (lanes I) is attained only with the nontransfected G3 cells but not with the EhLimA-silenced cells. No amplification is attained when the reverse transcription reaction is performed without reverse transcriptase (lanes II). Amplification of actin (lanes III) is attained with both the nontransfected G3 cells and with the EhLimA-silenced cells. (D) Western blot analysis of total cell lysates of G3 transfected trophozoites in which EhLimA is transcriptionally silenced. The anti-EhLimA antibodies do not detect any protein in these cells (lane 2), whereas the protein is detected in the nontransfected G3 cells expressing EhLimA (lane 1).
Transcriptional silencing of the EhLimA gene.
Transcriptional silencing of the EhLimA gene was achieved using a previously published technique generated in our lab (7). This technique allows us to induce the silencing of a second gene in the G3 substrain which is already silenced in the amoebapore gene (Ehap-a) and has an avirulent phenotype. Our aim in suppressing the expression of the EhLimA protein was to see whether this will have any effect on parasite growth or morphology. Silencing of EhLimA in the G3 strain was confirmed by an RT-PCR analysis that revealed an absence of EhLimA mRNA expression in the EhLimA-silenced amoeba (Fig. 3C). Similarly, in a Western blot analysis, no protein was detected in total lysates of the EhLimA-silenced amoeba (Fig. 3D). Silencing of EhLimA resulted in the absence of all four bands detected with the anti-EhLimA antibodies, indicating that all are derivatives or specific variants of EhLimA. Silencing was also confirmed in the localization studies (see below and see Fig. 4). The growth and morphology of the EhLimA-silenced amoeba did not appear to be affected.
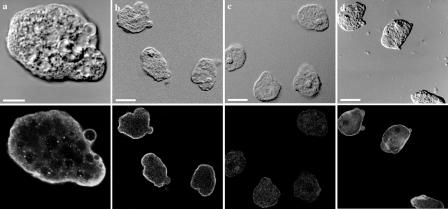
FIG. 4.
Immunofluorescence localization of EhLimA in whole parasites. Trophozoites were fixed and stained for immunofluorescence with anti-EhLimA antibodies followed by fluorescein isothiocyanate-conjugated anti-rabbit secondary antibodies and viewed under a confocal microscope. The differential interference contrast images are shown on the top, and the corresponding fluorescence images are shown on the bottom. EhLimA localizes mainly to the plasma membrane of cells (a and b). No staining was detected in the EhLimA-silenced G3 amoeba cells (c). Staining cells with phalloidin-TRITC reveals and enrichment of actin in the cell cortex and in cellular protrusions (d). Bars, 10 μm in panel a and 20 μm in panels b, c, and d.
Cellular localization of EhLimA.
The anti-EhLimA antibodies produced against recombinant EhLimA were also used to determine the cellular localization of EhLimA by indirect immunofluorescence. In almost all cells examined, EhLimA was highly enriched at the plasma membrane of the cells with some light staining in the cytoplasm (Fig. 4). A similar staining pattern was observed with both the parental HM-1:IMSS cells (Fig. 4a) as well as with the G3 substrain (Fig. 4b). The staining observed was very specific, as only a very faint signal was detected in the cytoplasm of the EhLimA-silenced amoeba (Fig. 4c). Amoeba cells were also stained with phalloidin that stains actin filaments (Fig. 4d). An enrichment of F-actin at the plasma membrane of cells resulting from staining of the underlying cortical actin was observed. F-actin was also highly enriched in cellular protrusions.
EhLimA associates with the cytoskeleton and interacts with actin.
Many LIM domain-containing proteins have been shown to associate with the actin cytoskeleton (31), including the Dictyostelium homologs DdLimE (45, 48) and LimD (30) which were found to localize to the cell cortex. Localization of EhLimA to the plasma membrane led us to examine whether EhLimA associates with the underlying actin cytoskeleton. The presence of EhLimA in cytoskeletal fractions was first examined. Total cell lysates prepared with the addition of Triton X-100 were subjected to ultracentrifugation, resulting in a pellet fraction containing the Triton X-100-insoluble cytoskeleton and a supernatant fraction containing the soluble proteins. Proteins from these fractions were separated by SDS-PAGE followed by Western blotting with anti-EhLimA antibodies (Fig. 5). The results show that a significant portion of EhLimA is found in the Triton X-100-insoluble cytoskeleton fraction. The majority of actin was detected in this fraction, also confirming the specificity of this fraction. These results indicate that EhLimA is associated with the cytoskeleton.
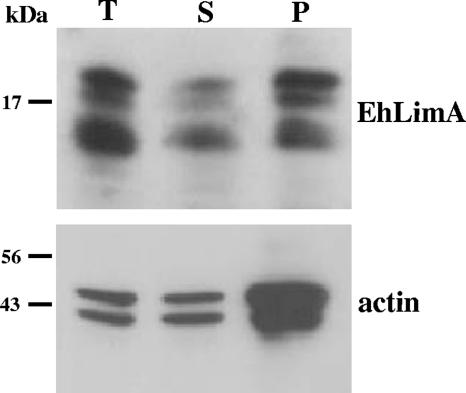
FIG. 5.
EhLimA associates with the cytoskeleton. Total cell lysates (T) prepared with the addition of Triton X-100 were subjected to ultracentrifugation resulting in a pellet fraction (P) containing the Triton X-100-insoluble cytoskeleton and a supernatant fraction (S) containing soluble proteins. A Western blot analysis of these fractions using anti-EhLimA antibodies shows that a significant portion of EhLimA is found in the Triton X-100-insoluble cytoskeleton fraction. Monoclonal antiactin antibodies reveal that the majority of actin is detected in this fraction as well.
The cytoskeleton association of EhLimA was further examined by assaying whether actin binding is involved. A construct was prepared that encoded full-length EhLimA (amino acid residues 1 to 145) tagged at the N terminus with the 24 nucleotides (8 amino acids) corresponding to the FLAG epitope (FLAG-EhLimA; Fig. 6A, panel a). This construct was cloned into an amoeba overexpression vector under the control of the Ehactin gene promoter and transfected into amoeba cells. Protein expression of FLAG-EhLimA was confirmed by Western blotting with both anti-EhLimA antibodies as well as anti-FLAG monoclonal antibodies (Fig. 6B). Interestingly, in FLAG-EhLimA-overexpressing cells, the anti-EhLimA antibodies detected in addition to the bands representing endogenous EhLimA a slightly larger band probably representing the FLAG-EhLimA fusion protein, while the anti-FLAG antibodies recognized this larger band but no additional bands. The FLAG-EhLimA fusion protein was immunoprecipitated from cells using anti-FLAG monoclonal antibodies covalently attached to agarose. Purified FLAG-EhLimA was analyzed by SDS-PAGE, followed by total protein staining of the gel (Fig. 6C, top gel). As can be seen, the amount of purified protein detected in the gel was in direct correlation to the amount of total cell lysate used in the assay. Immunoprecipitated protein products were analyzed in a Western blot using antiactin antibodies in order to determine whether actin is coimmunoprecipitated together with EhLimA (Fig. 6C, bottom gel). Indeed, actin was detected in the lanes corresponding to cells overexpressing FLAG-EhLimA, and the amount of actin detected was directly related to the amount of purified FLAG-EhLimA protein in each lane. No actin was detected in the lane corresponding to wild-type cells. These results clearly show that EhLimA associates with actin and that this association is specific. Interestingly, we noticed that while the antiactin antibodies identify two bands in total cell lysates, only the upper band appears in the lanes containing the purified FLAG-EhLimA protein. The lower band may represent a degraded form of actin that does not associate with EhLimA. A smaller actin fragment was recently identified in a two-dimensional gel electrophoresis analysis of the E. histolytica proteome (34). Taken together, it is reasonable to assume that plasma membrane localization of EhLimA is due at least in part to an association of EhLimA with the underlying cortical actin cytoskeleton.
FIG. 6.
(A) Schematic representation of constructs encoding full-length and truncated FLAG-tagged EhLimA. (a) N-terminus-tagged full-length EhLimA (FLAG-EhLimA); (b) N-terminus-tagged EhLimA lacking half of the N terminus LIM domain (N-TRUNC); (c) N-terminus-tagged EhLimA lacking the glutamic acid-rich C terminus (C-TRUNC); (d) C-terminus-tagged full-length EhLimA (C-FLAG). (B) Protein expression of FLAG-EhLimA is confirmed in a Western blot analysis using anti-EhLimA antibodies (lanes I) and anti-FLAG monoclonal antibodies (lanes II). Lane 1 and 2 contain total cell lysates of wild-type cells and FLAG-EhLimA-overexpressing cells, respectively. (C) Different amounts of total cell lysates of FLAG-EhLimA-overexpressing cells (lanes 1 to 3) and wild-type cells (lane 5) were incubated with anti-FLAG monoclonal antibodies covalently attached to agarose. Immunoprecipitated FLAG-EhLimA was analyzed by gel electrophoresis followed by total protein staining of the gel (top gel). Coimmunoprecipitation of actin was analyzed in a Western blot of immunoprecipitated protein products using antiactin antibodies (bottom gel). Lane 1, 250 μg total lysate; lane 2, 100 μg total lysate; lane 3, 50 μg total lysate; lane 5, 500 μg total lysate. Protein amounts are confirmed in lanes 4 and 6 for FLAG-EhLimA-overexpressing cells and wild-type cells, respectively, each containing 10 μg of total lysate.
Actin binding is dependent on the N terminus but not the C terminus of EhLimA.
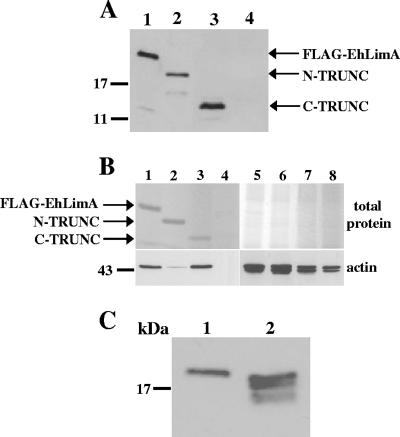
In order to identify the region of the protein involved in actin binding, two constructs were prepared encoding EhLimA truncated at either the N terminus (amino acid residues 31 to 145) or C terminus (amino acid residues 1 to 100) tagged at the N terminus with the FLAG epitope. The N-terminus-truncated and FLAG-tagged EhLimA (N-TRUNC) is lacking half of the LIM domain (Fig. 6A, panel b), while the C-terminus-truncated FLAG-tagged EhLimA (C-TRUNC) is lacking the glutamic acid-rich region of the protein (Fig. 6A, panel c). Both constructs were cloned into an amoeba overexpression vector under the control of the Ehactin gene promoter and transfected into amoeba cells. Protein expression of the truncated FLAG-tagged EhLimA proteins was verified by Western blotting using anti-FLAG monoclonal antibodies (Fig. 7A). The truncated proteins were immunoprecipitated using anti-FLAG antibodies covalently attached to agarose. PAGE of the immunoprecipitated protein products revealed nice amounts of both truncated proteins (Fig. 7B, top gel). The ability of the truncated EhLimA proteins to associate with actin was verified in a Western blot analysis of immunoprecipitated protein products using antiactin antibodies (Fig. 7B, bottom gel). While the intensity of the band detected in the lane containing purified C-TRUNC was similar to that of the band detected in the control lane containing the purified full-length FLAG-EhLimA fusion protein, a very light band was detected in the lane containing purified N-TRUNC. These results show that the association between EhLimA and actin is dependent on the N terminus of EhLimA, which contains the LIM domain, and that removal of even half of the LIM domain significantly inhibits the ability of EhLimA to associate with actin. The C terminus of EhLimA, on the other hand, does not appear to be involved in actin binding, as removal of this region of the protein does not affect the ability of the protein to associate with actin.
FIG. 7.
(A) Expression of N-terminus- and C-terminus-truncated FLAG-tagged EhLimA. Protein expression of N-terminus-truncated FLAG-tagged EhLimA (N-TRUNC, lane 2) and C-terminus-truncated FLAG-tagged EhLimA (C-TRUNC, lane 3) is confirmed in a Western blot analysis with anti-FLAG antibodies. In lane 1, expression of the full-length FLAG-EhLimA fusion protein is shown. Lane 4 contains a total cell lysate of wild-type cells. (B) Immunoprecipitation and actin binding of N-terminus- and C-terminus-truncated FLAG-tagged EhLimA. Total cell lysates of cells overexpressing FLAG- EhLimA (lane 1), N-TRUNC (lane 2), or C-TRUNC (lane 3) and wild-type cells (lane 4) were incubated with anti-FLAG monoclonal antibodies covalently attached to agarose. Immunoprecipitated proteins were analyzed by gel electrophoresis, followed by total protein staining of the gel (top gel). A Western blot analysis of immunoprecipitated protein products using antiactin antibodies reveals that N-TRUNC has almost completely lost the ability to bind actin (bottom gel). Protein amounts are confirmed in lanes 5 to 8 for lanes 1 to 4, respectively, each containing 10 μg of total lysate. (C) N-terminal cleavage of EhLimA. Anti-FLAG antibodies detect four bands in a Western blot of total cell lysates of cells overexpressing FLAG-tagged full-length EhLimA in which the FLAG epitope is located at the C terminus of the protein (C-FLAG, lane 2) as opposed to only one band that is detected in total cell lysates of the N-terminus-tagged FLAG-EhLimA-overexpressing cells (lane 1).
N-terminal cleavage of EhLimA.
The observation that the anti-EhLimA antibodies detect four bands in Western blots of amoeba total cell lysates suggested that this protein undergoes some type of cleavage or modification resulting in a number of protein variants. Though we considered the possibility that these bands were the result of unspecific degradation, the gel migration pattern of the bands was highly reproducible and was observed in all Western blots analyzed. In addition, if there was unspecific degradation, it would most likely also affect other proteins analyzed and this was not the case. As shown in Fig. 6B, in FLAG-EhLimA-overexpressing cells, the anti- EhLimA antibodies detect in addition to these bands, a slightly larger band representing the FLAG-EhLimA fusion protein. However, the anti-FLAG antibodies recognize only the FLAG-EhLimA protein in these cells. The inability of the anti-FLAG antibodies to recognize any additional bands suggests that these bands are the result of N-terminal cleavage of EhLimA. Such cleavage would result in removal of the FLAG epitope that is located at the N terminus of the protein, and N-terminally cleaved variants of EhLimA would not be detected by the anti-FLAG antibodies. N-terminal cleavage of EhLimA was further confirmed by overexpressing a construct encoding full-length EhLimA tagged at the C terminus with the FLAG epitope (C-FLAG; Fig. 6A, panel d). In lysates of cells overexpressing C-terminus-tagged full-length EhLimA, four bands were detected by the anti-FLAG antibodies as opposed to only one band that is detected in lysates of cells overexpressing the N-terminus-tagged FLAG-EhLimA fusion protein (Fig. 7C). Taken together, these data indicate that EhLimA undergoes N-terminal cleavage.
Evidence that a portion of EhLimA associates with lipid rafts in the parasite membrane.
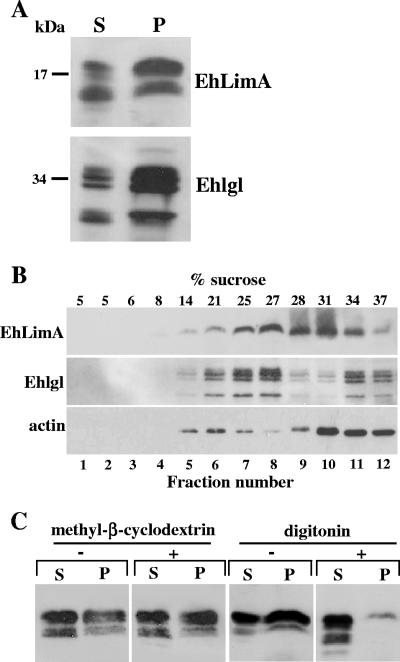
Lipid rafts are membrane domains that have been found to play a role in various cellular functions, including membrane trafficking, endocytosis, secretion, and signal transduction (13, 52, 53). Recently, raft-like microdomains were identified in E. histolytica and were found to be involved in pinocytosis and adhesion (33). Insolubility in Triton X-100, which is a property common to cytoskeleton-associated proteins, can also serve as an indication that the protein associates with lipid rafts (25, 49) that are insoluble in Triton X-100 due to their high cholesterol and sphingolipid content (14, 52). Insolubility of EhLimA in Triton X-100 was observed following ultracentrifugation as shown in Fig. 5, or when cell lysates were fractionated at lower speed centrifugation (13,000 × g) into supernatant (soluble) and pellet (insoluble) fractions (Fig. 8A). In this case also, a significant portion of EhLimA was found in the Triton X-100-insoluble pellet fraction. Interestingly, a similar distribution of the amoebic Gal-lectin light subunit (Ehlgl), which was reported to be present in raft-like fractions (33), was observed in these fractions as well.
FIG. 8.
A portion of EhLimA associates with lipid rafts. (A) Total cell lysates prepared with the addition of cold Triton X-100 were subjected to centrifugation resulting in soluble (S) and insoluble (P) fractions. A Western blot analysis using anti-EhLimA antibodies shows that a significant portion of EhLimA is found in the Triton X-100-insoluble fraction. A similar protein distribution of the amoebic Gal-lectin light subunit (Ehlgl) was detected using polyclonal anti-Ehlgl antibodies. (B) Total cell lysates prepared with cold Triton X-100 were subjected to sucrose gradient flotation. Twelve fractions were collected from the top of the gradient and proteins were precipitated with trichloroacetic acid. Following SDS-PAGE and immunoblotting, the distribution of EhLimA, Ehlgl, and actin in the fractions was analyzed. A portion of EhLimA as well as of Ehlgl and actin was detected in the lower-density regions of the gradient. The presence of EhLimA in the lower-density regions was observed in three independent experiments. The percentage of sucrose in each fraction is indicated. (C) Depletion of cholesterol. Cells were incubated for 30 min with either digitonin or methyl-β-cyclodextrin on ice followed by lysis in Triton X-100 and centrifugation. Western blotting reveals that the distribution of EhLimA between soluble (S) and insoluble (P) fractions was greatly affected by digitonin but not by methyl-β-cyclodextrin.
The association of EhLimA with lipid rafts was assayed by sucrose gradient centrifugation, a method commonly used for isolating lipid rafts (13, 53). Total cell lysates prepared with cold Triton X-100 were loaded at the bottom of a sucrose gradient and subjected to sucrose gradient flotation. Fractions were collected from the top of the gradient, and the distribution of EhLimA as well as of Ehlgl and actin was analyzed in the collected fractions by SDS-PAGE followed by immunoblotting (Fig. 8B). Following sucrose gradient flotation, a portion of EhLimA was detected in the lower-density regions of the gradient. EhLimA was also detected in other regions, including the high-density regions of the gradient. This was expected, since we have shown that EhLimA associates with the actin cytoskeleton that itself distributes to the high-density regions of the gradient, as seen here by the predominance of actin in these fractions. Ehlgl, which as already mentioned has been shown to interact with lipid microdomains in E. histolytica (33), was used as a marker for raft-like domains and was indeed found to be enriched in the lower-density regions of the gradient, though it also appeared in the higher-density regions. Results obtained in three independent experiments revealed some differences in the distribution of EhLimA, but in all experiments, a portion of EhLimA was found in association with the lower-density regions of the gradient.
Depletion or sequestration of cholesterol from the membrane many times renders raft-associated proteins detergent soluble as a result of raft disruption. The cholesterol-depleting agent methyl-β-cyclodextrin has been shown to increase the detergent solubility of various raft-associated proteins (32, 47). Digitonin that disrupts rafts by sequestering membrane sterols has resulted in increased solubility of a protein associated with the Triton X-100-insoluble floating fraction in Dictyostelium (26). Amoeba cells were incubated on ice with either methyl-β-cyclodextrin or digitonin in order to determine whether the Triton X-100 insolubility of EhLimA is dependent on the presence of cholesterol. The cells were then lysed with Triton X-100 and fractionated by centrifugation into soluble and insoluble fractions (Fig. 8C). Analysis of these fractions revealed that treatment of cells with methyl-β-cyclodextrin did not appear to have any effect on the solubility of EhLimA, as the distribution of the protein between soluble and insoluble fractions was not affected. Similar results were obtained when methyl-β-cyclodextrin-treated cells were incubated at 37°C instead of on ice (not shown). When cells were treated with digitonin, however, the solubility of EhLimA increased dramatically, as most of the protein was detected in the supernatant and only a very small amount was detected in the insoluble pellet. The almost complete solubilization of EhLimA by digitonin indicates that the Triton X-100 insolubility of EhLimA is due to an association of EhLimA with cholesterol-rich lipid rafts in the membrane. It has been reported that there are cases where methyl-β-cyclodextrin does not solubilize raft-associated proteins (44). The reason for the lack of solubilization of EhLimA by methyl-β-cyclodextrin in E. histolytica trophozoites may also be due to differences in membrane lipid composition of this parasite.
DISCUSSION
Various cellular activities, including motility and chemotaxis, adhesion, endocytosis, phagocytosis, and cytokinesis, are dependent on the cytoskeleton of the cell. In order to invade and survive in host tissues, E. histolytica trophozoites rely on a highly dynamic cytoskeleton to perform such processes as phagocytosis, motility, or adhesion that are required for the pathogenesis of E. histolytica (23). Various components of the cytoskeleton have been identified in E. histolytica (37), and a number of proteins have been shown to bind actin, including myosin IB (56), calcium binding protein 1 (46), and the actin binding protein ABP-120 (55). In a wide variety of eukaryotic organisms, LIM domain-containing proteins have been shown to interact directly or indirectly with the actin cytoskeleton (27, 31). The recent completion of the E. histolytica Genome Project (35) revealed the presence of several LIM domain-containing proteins present in the E. histolytica genome. Of these, 12 are annotated as LIM domain proteins or proteins containing LIM domains, but the total number of LIM domain-containing proteins is apparently higher, as some LIM domain-containing proteins have been annotated as proteins to which they are homologous.
Here we have characterized the first LIM domain-containing protein of E. histolytica which we have termed EhLimA. EhLimA contains a single LIM domain at its N terminus and shares the highest degree of homology with the Dictyostelium DdLimE LIM domain protein. DdLimE localizes to the cell cortex and regulates cell motility and cytokinesis (45, 48). We found that EhLimA associates with the cytoskeleton and that cytoskeleton association involves actin binding. This is shown by the detection of a significant portion of EhLimA in the actin-enriched cytoskeleton fraction of cells. In addition, immunoprecipitation of a FLAG-EhLimA fusion protein from cells resulted in coimmunoprecipitation of actin, indicating that EhLimA is associated with actin. Our results show that the N-terminal LIM domain of EhLimA is necessary for actin binding, which is consistent with the role of the LIM domain in mediating protein-protein interactions. We found that removal of even half of the LIM domain from the protein almost completely prevented actin binding. On the other hand, we did not find that the glutamic acid-rich C terminus plays a role in actin binding as removal of this region of the protein had no effect on actin binding.
Our results also suggest a possible association between EhLimA and raft-like microdomains in the E. histolytica plasma membrane. First of all, a significant portion of EhLimA was not solubilized by cold Triton X-100. Triton X-100 insolubility is a property characteristic of cytoskeleton-associated proteins but may also indicate that the protein associates with lipid rafts (25, 49). In the case of EhLimA, Triton X-100 insolubility may be the result of associations with both the cytoskeleton and lipid rafts. Association of EhLimA with lipid rafts was shown by the distribution of a portion of EhLimA to the lower-density regions of a sucrose gradient. Since raft-like microdomains have only recently been identified in E. histolytica (33), at present there is no well-accepted raft marker for this organism. We used the light subunit of the amoebic Gal-lectin (Ehlgl) that was shown to interact with lipid microdomains in E. histolytica (33) and found that it was indeed enriched in the lower-density regions of the sucrose gradient.
Increased solubility of many raft-associated proteins is achieved by cholesterol depletion (6, 15, 32, 47). We found that treatment of cells with the cholesterol-depleting agent methyl-β-cyclodextrin had no effect on the solubility of EhLimA, while the cholesterol-complexing agent digitonin resulted in almost complete solubilization of EhLimA. In a previous study (50), it was reported that methyl-β-cyclodextrin was effective in disassociating proteins from detergent-resistant membranes only when used on homogenates of MDCK cells but not when used on intact cells. Interestingly, in another study (1), the amount of proaerolysin, an inactive precursor of the bacterial toxin aerolysin, in the detergent-insoluble pellet and in the solubilized fraction of BHK cells was only slightly affected by methyl-β-cyclodextrin, while saponin treatment resulted in redistribution of most of the protein to the detergent-soluble fraction. These findings indicate that the ability of cholesterol-affecting agents to disrupt rafts may be highly dependent on the state of the cells during treatment with these agents and that not all rafts whose integrity depends on cholesterol will be equally affected by different cholesterol-affecting agents. In addition, variations in the lipid composition of different rafts may affect the protein solubilization properties of cholesterol-depleting agents. Taken together, the results obtained in the sucrose gradient flotation assay together with the observation that the cholesterol-sequestering agent digitonin almost completely abolishes the Triton X-100 insolubility of EhLimA indicate that in addition to being associated with the actin cytoskeleton, a portion of EhLimA may also associate with raft-like microdomains in the plasma membrane.
The observed localization of EhLimA to the plasma membrane of cells can be attributed to both an association of EhLimA with the underlying cortical actin cytoskeleton and to some degree with membrane rafts. While association of EhLimA with the cytoskeleton is mediated through actin binding and involves the N-terminal LIM domain of EhLimA, the mechanism responsible for the association of EhLimA with rafts is not as clear. EhLimA does not possess any known structural features, such as a glycosylphosphatidylinositol anchor, acyl tails, or a transmembrane domain, that could account for raft association (52). We have not established whether the interaction between EhLimA and actin takes place directly or indirectly, and it is possible that their interaction occurs indirectly with an additional protein or proteins mediating the interaction between the LIM domain of EhLimA and both actin and lipid rafts. Such proteins may include actin binding proteins that have the ability to bind phosphoinositide lipids, such as phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] (51), and are thought to link the actin cytoskeleton with the plasma membrane and in some cases with rafts. Examples include ERM (ezrin/radixin/moesin) proteins that can bind to phosphoinositides (41) and are considered mediators of interactions between the actin cytoskeleton and plasma membrane (2, 11, 12). Gelsolin, an actin-capping and -severing protein that binds to phosphoinositides has been shown to associate with rafts (22). A different example involves the membrane raft protein PAG, which was found to bind to the cytoplasmic adaptor protein EBP50 (10) that is known to interact with the ERM-family proteins that bind to the actin cytoskeleton. In this case, a number of proteins participate in connecting membrane rafts to the actin cytoskeleton. Similarly, our results suggest that EhLimA may be part of the molecular system connecting the actin cytoskeleton to membrane rafts. Since our results show that EhLimA is cleaved at the N terminus, N-terminal cleavage may serve as a regulatory mechanism rendering EhLimA unable to associate with protein partners and thereby affecting its ability to interact with the cytoskeleton and membrane rafts.
In agreement with the role of the LIM domain as a protein binding interface, it is conceivable that the LIM domain of EhLimA serves as a platform for the formation of a multiprotein complex that includes proteins possessing both actin binding capabilities and lipid binding capabilities and that association of EhLimA with actin and lipid rafts takes place as part of this multiprotein complex. While identification of such proteins will be the subject of future work, preliminary results have identified the glycolytic enzyme enolase as a putative binding partner of EhLimA. Though generally found in the cytoplasm, a number of studies have identified α-enolase on the surfaces of many cell types (43). Furthermore, in a recent study, a small fraction of enolase from the malarial parasite Plasmodium yoeli was found to be associated with cell membranes and cytoskeletal elements (42). This makes enolase an interesting candidate for mediating the interactions between EhLimA and the cytoskeleton and/or lipid rafts.
Our results also show that silencing or overexpression of EhLimA did not cause any significant effect on the growth or morphology of the parasite. Since the E. histolytica genome contains several putative LIM proteins (35), it is possible that in the absence of EhLimA, other LIM proteins compensate for its function. In addition, because the amoeba substrain G3 in which the silencing of a second gene can be induced is already avirulent, it was not possible to investigate whether EhLimA plays any role in parasite virulence.
Acknowledgments
This investigation was supported by a grant from the Drake Family Foundation. The stipend of N.W. was funded by a grant from Henry H. Meyer, Jr.
We thank Talia Miron for her help with the purification of the anti-EhLimA antibodies. We also thank Rivka Bracha and Yael Nuchamowitz for their suggestions and assistance.
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Abrami, L., and F. G. van Der Goot. 1999. Plasma membrane microdomains act as concentration platforms to facilitate intoxication by aerolysin. J. Cell Biol. 147:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Algrain, M., O. Turunen, A. Vaheri, D. Louvard, and M. Arpin. 1993. Ezrin contains cytoskeleton and membrane binding domains accounting for its proposed role as a membrane-cytoskeletal linker. J. Cell Biol. 120:129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ankri, S., F. Padilla-Vaca, T. Stolarsky, L. Koole, U. Katz, and D. Mirelman. 1999. Antisense inhibition of expression of the light subunit (35 kDa) of the Gal/GalNac lectin complex inhibits Entamoeba histolytica virulence. Mol. Microbiol. 33:327-337. [DOI] [PubMed] [Google Scholar]
- 4.Bach, I. 2000. The LIM domain: regulation by association. Mech. Dev. 91:5-17. [DOI] [PubMed] [Google Scholar]
- 5.Baltz, R., C. Domon, D. T. Pillay, and A. Steinmetz. 1992. Characterization of a pollen-specific cDNA from sunflower encoding a zinc finger protein. Plant J. 2:713-721. [PubMed] [Google Scholar]
- 6.Becher, A., J. H. White, and R. A. McIlhinney. 2001. The γ-aminobutyric acid receptor B, but not the metabotropic glutamate receptor type-1, associates with lipid rafts in the rat cerebellum. J. Neurochem. 79:787-795. [DOI] [PubMed] [Google Scholar]
- 7.Bracha, R., Y. Nuchamowitz, M. Anbar, and D. Mirelman. 2006. Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog. 2:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bracha, R., Y. Nuchamowitz, and D. Mirelman. 2003. Transcriptional silencing of an amoebapore gene in Entamoeba histolytica: molecular analysis and effect on pathogenicity. Eukaryot. Cell 2:295-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Brdickova, N., T. Brdicka, L. Andera, J. Spicka, P. Angelisova, S. L. Milgram, and V. Horejsi. 2001. Interaction between two adapter proteins, PAG and EBP50: a possible link between membrane rafts and actin cytoskeleton. FEBS Lett. 507:133-136. [DOI] [PubMed] [Google Scholar]
- 11.Bretscher, A. 1999. Regulation of cortical structure by the ezrin-radixin-moesin protein family. Curr. Opin. Cell Biol. 11:109-116. [DOI] [PubMed] [Google Scholar]
- 12.Bretscher, A., D. Reczek, and M. Berryman. 1997. Ezrin: a protein requiring conformational activation to link microfilaments to the plasma membrane in the assembly of cell surface structures. J. Cell Sci. 110:3011-3018. [DOI] [PubMed] [Google Scholar]
- 13.Brown, D. A., and E. London. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14:111-136. [DOI] [PubMed] [Google Scholar]
- 14.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. [DOI] [PubMed] [Google Scholar]
- 15.Cerneus, D. P., E. Ueffing, G. Posthuma, G. J. Strous, and A. van der Ende. 1993. Detergent insolubility of alkaline phosphatase during biosynthetic transport and endocytosis. Role of cholesterol. J. Biol. Chem. 268:3150-3155. [PubMed] [Google Scholar]
- 16.Chien, S., C. Y. Chung, S. Sukumaran, N. Osborne, S. Lee, C. Ellsworth, J. G. McNally, and R. A. Firtel. 2000. The Dictyostelium LIM domain-containing protein LIM2 is essential for proper chemotaxis and morphogenesis. Mol. Biol. Cell 11:1275-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis, P. H., X. Zhang, J. Guo, R. R. Townsend, and S. L. Stanley, Jr. 2006. Comparative proteomic analysis of two Entamoeba histolytica strains with different virulence phenotypes identifies peroxiredoxin as an important component of amoebic virulence. Mol. Microbiol. 61:1523-1532. [DOI] [PubMed] [Google Scholar]
- 18.Dawid, I. B., J. J. Breen, and R. Toyama. 1998. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14:156-162. [DOI] [PubMed] [Google Scholar]
- 19.Diamond, L. S., D. R. Harlow, and C. C. Cunnick. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431-432. [DOI] [PubMed] [Google Scholar]
- 20.Eliasson, A., N. Gass, C. Mundel, R. Baltz, R. Krauter, J. L. Evrard, and A. Steinmetz. 2000. Molecular and expression analysis of a LIM protein gene family from flowering plants. Mol. Gen. Genet. 264:257-267. [DOI] [PubMed] [Google Scholar]
- 21.Freyd, G., S. K. Kim, and H. R. Horvitz. 1990. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature 344:876-879. [DOI] [PubMed] [Google Scholar]
- 22.Funatsu, N., H. Kumanogoh, Y. Sokawa, and S. Maekawa. 2000. Identification of gelsolin as an actin regulatory component in a triton insoluble low density fraction (raft) of newborn bovine brain. Neurosci. Res. 36:311-317. [DOI] [PubMed] [Google Scholar]
- 23.Guillen, N. 1996. Role of signalling and cytoskeletal rearrangements in the pathogenesis of Entamoeba histolytica. Trends Microbiol. 4:191-197. [DOI] [PubMed] [Google Scholar]
- 24.Hamann, L., R. Nickel, and E. Tannich. 1995. Transfection and continuous expression of heterologous genes in the protozoan parasite Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 92:8975-8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harder, T., and K. Simons. 1997. Caveolae, DIGs, and the dynamics of sphingolipid-cholesterol microdomains. Curr. Opin. Cell Biol. 9:534-542. [DOI] [PubMed] [Google Scholar]
- 26.Harris, T. J., D. E. Awrey, B. J. Cox, A. Ravandi, A. Tsang, and C. H. Siu. 2001. Involvement of a triton-insoluble floating fraction in Dictyostelium cell-cell adhesion. J. Biol. Chem. 276:18640-18648. [DOI] [PubMed] [Google Scholar]
- 27.Kadrmas, J. L., and M. C. Beckerle. 2004. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5:920-931. [DOI] [PubMed] [Google Scholar]
- 28.Karlsson, O., S. Thor, T. Norberg, H. Ohlsson, and T. Edlund. 1990. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature 344:879-882. [DOI] [PubMed] [Google Scholar]
- 29.Katz, U., S. Ankri, T. Stolarsky, Y. Nuchamowitz, and D. Mirelman. 2002. Entamoeba histolytica expressing a dominant negative N-truncated light subunit of its gal-lectin are less virulent. Mol. Biol. Cell 13:4256-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khurana, B., T. Khurana, N. Khaire, and A. A. Noegel. 2002. Functions of LIM proteins in cell polarity and chemotactic motility. EMBO J. 21:5331-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khurana, T., B. Khurana, and A. A. Noegel. 2002. LIM proteins: association with the actin cytoskeleton. Protoplasma 219:1-12. [DOI] [PubMed] [Google Scholar]
- 32.Lafont, F., S. Lecat, P. Verkade, and K. Simons. 1998. Annexin XIIIb associates with lipid microdomains to function in apical delivery. J. Cell Biol. 142:1413-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laughlin, R. C., G. C. McGugan, R. R. Powell, B. H. Welter, and L. A. Temesvari. 2004. Involvement of raft-like plasma membrane domains of Entamoeba histolytica in pinocytosis and adhesion. Infect. Immun. 72:5349-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leitsch, D., C. Radauer, K. Paschinger, I. B. Wilson, H. Breiteneder, O. Scheiner, and M. Duchene. 2005. Entamoeba histolytica: analysis of the trophozoite proteome by two-dimensional polyacrylamide gel electrophoresis. Exp. Parasitol. 110:191-195. [DOI] [PubMed] [Google Scholar]
- 35.Loftus, B., I. Anderson, R. Davies, U. C. Alsmark, J. Samuelson, P. Amedeo, P. Roncaglia, M. Berriman, R. P. Hirt, B. J. Mann, T. Nozaki, B. Suh, M. Pop, M. Duchene, J. Ackers, E. Tannich, M. Leippe, M. Hofer, I. Bruchhaus, U. Willhoeft, A. Bhattacharya, T. Chillingworth, C. Churcher, Z. Hance, B. Harris, D. Harris, K. Jagels, S. Moule, K. Mungall, D. Ormond, R. Squares, S. Whitehead, M. A. Quail, E. Rabbinowitsch, H. Norbertczak, C. Price, Z. Wang, N. Guillen, C. Gilchrist, S. E. Stroup, S. Bhattacharya, A. Lohia, P. G. Foster, T. Sicheritz-Ponten, C. Weber, U. Singh, C. Mukherjee, N. M. El-Sayed, W. A. Petri, Jr., C. G. Clark, T. M. Embley, B. Barrell, C. M. Fraser, and N. Hall. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865-868. [DOI] [PubMed] [Google Scholar]
- 36.Marion, S., C. Laurent, and N. Guillen. 2005. Signalization and cytoskeleton activity through myosin IB during the early steps of phagocytosis in Entamoeba histolytica: a proteomic approach. Cell. Microbiol. 7:1504-1518. [DOI] [PubMed] [Google Scholar]
- 37.Meza, I., P. Talamas-Rohana, and M. A. Vargas. 2006. The cytoskeleton of Entamoeba histolytica: structure, function, and regulation by signaling pathways. Arch. Med. Res. 37:234-243. [DOI] [PubMed] [Google Scholar]
- 38.Moshitch-Moshkovitch, S., T. Stolarsky, D. Mirelman, and R. N. Alon. 1996. Stable episomal transfection and gene expression in Entamoeba dispar. Mol. Biochem. Parasitol. 83:257-261. [DOI] [PubMed] [Google Scholar]
- 39.Muller, L., G. Xu, R. Wells, C. P. Hollenberg, and W. Piepersberg. 1994. LRG1 is expressed during sporulation in Saccharomyces cerevisiae and contains motifs similar to LIM and rho/racGAP domains. Nucleic Acids Res. 22:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mundel, C., R. Baltz, A. Eliasson, R. Bronner, N. Grass, R. Krauter, J. L. Evrard, and A. Steinmetz. 2000. A LIM-domain protein from sunflower is localized to the cytoplasm and/or nucleus in a wide variety of tissues and is associated with the phragmoplast in dividing cells. Plant Mol. Biol. 42:291-302. [DOI] [PubMed] [Google Scholar]
- 41.Niggli, V., C. Andreoli, C. Roy, and P. Mangeat. 1995. Identification of a phosphatidylinositol-4,5-bisphosphate-binding domain in the N-terminal region of ezrin. FEBS Lett. 376:172-176. [DOI] [PubMed] [Google Scholar]
- 42.Pal-Bhowmick, I., H. K. Vora, and G. K. Jarori. 2007. Sub-cellular localization and post-translational modifications of the Plasmodium yoelii enolase suggest moonlighting functions. Malaria J. 6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pancholi, V. 2001. Multifunctional α-enolase: its role in diseases. Cell. Mol. Life Sci. 58:902-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pike, L. J. 2004. Lipid rafts: heterogeneity on the high seas. Biochem. J. 378:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prassler, J., A. Murr, S. Stocker, J. Faix, J. Murphy, and G. Marriott. 1998. DdLIM is a cytoskeleton-associated protein involved in the protrusion of lamellipodia in Dictyostelium. Mol. Biol. Cell 9:545-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sahoo, N., E. Labruyere, S. Bhattacharya, P. Sen, N. Guillen, and A. Bhattacharya. 2004. Calcium binding protein 1 of the protozoan parasite Entamoeba histolytica interacts with actin and is involved in cytoskeleton dynamics. J. Cell Sci. 117:3625-3634. [DOI] [PubMed] [Google Scholar]
- 47.Scheiffele, P., M. G. Roth, and K. Simons. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider, N., I. Weber, J. Faix, J. Prassler, A. Muller-Taubenberger, J. Kohler, E. Burghardt, G. Gerisch, and G. Marriott. 2003. A Lim protein involved in the progression of cytokinesis and regulation of the mitotic spindle. Cell Motil. Cytoskelet. 56:130-139. [DOI] [PubMed] [Google Scholar]
- 49.Schroeder, R., E. London, and D. Brown. 1994. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc. Natl. Acad. Sci. USA 91:12130-12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuck, S., M. Honsho, K. Ekroos, A. Shevchenko, and K. Simons. 2003. Resistance of cell membranes to different detergents. Proc. Natl. Acad. Sci. USA 100:5795-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sechi, A. S., and J. Wehland. 2000. The actin cytoskeleton and plasma membrane connection: PtdIns(4,5)P2 influences cytoskeletal protein activity at the plasma membrane. J. Cell Sci. 113:3685-3695. [DOI] [PubMed] [Google Scholar]
- 52.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 53.Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1:31-39. [DOI] [PubMed] [Google Scholar]
- 54.Taira, M., J. L. Evrard, A. Steinmetz, and I. B. Dawid. 1995. Classification of LIM proteins. Trends Genet. 11:431-432. [DOI] [PubMed] [Google Scholar]
- 55.Vargas, M., P. Sansonetti, and N. Guillen. 1996. Identification and cellular localization of the actin-binding protein ABP-120 from Entamoeba histolytica. Mol. Microbiol. 22:849-857. [DOI] [PubMed] [Google Scholar]
- 56.Vargas, M., H. Voigt, P. Sansonetti, and N. Guillen. 1997. Molecular characterization of myosin IB from the lower eukaryote Entamoeba histolytica, a human parasite. Mol. Biochem. Parasitol. 86:61-73. [PubMed] [Google Scholar]
- 57.Way, J. C., and M. Chalfie. 1988. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell 54:5-16. [DOI] [PubMed] [Google Scholar]