Abstract
Bioactive small molecules are critical in Aspergillus species during their development and interaction with other organisms. Genes dedicated to their production are encoded in clusters that can be located throughout the genome. We show that deletion of hdaA, encoding an Aspergillus nidulans histone deacetylase (HDAC), causes transcriptional activation of two telomere-proximal gene clusters—and subsequent increased levels of the corresponding molecules (toxin and antibiotic)—but not of a telomere-distal cluster. Introduction of two additional HDAC mutant alleles in a ΔhdaA background had minimal effects on expression of the two HdaA-regulated clusters. Treatment of other fungal genera with HDAC inhibitors resulted in overproduction of several metabolites, suggesting a conserved mechanism of HDAC repression of some secondary-metabolite gene clusters. Chromatin regulation of small-molecule gene clusters may enable filamentous fungi to successfully exploit environmental resources by modifying chemical diversity.
A distinguishing characteristic of filamentous fungi is their ability to produce a wide variety of small molecules that aid in their survival and pathogenicity. These include compounds, such as pigments, that play a role in virulence and protect the fungus from environmental damage, as well as toxins that kill host tissues or hinder competition from other organisms. These secondary metabolites (SM) (27) can also impact humans in both beneficial and detrimental ways. Many widely used pharmaceuticals are natural products of fungi, as are some of the most potent carcinogens yet identified. Genetic studies, augmented by analysis of whole genome sequences, have revealed that most fungal SM biosynthetic genes are found in compact clusters functioning as individual genetic loci (27). The genus Aspergillus, whose members include toxin-producing pathogens (Aspergillus flavus and A. fumigatus) and pharmaceutical-producing species (A. nidulans and A. terreus), is renowned for prodigious metabolite production and serves as the model for natural-product exploration. Detailed comparison of the genomes of several aspergilli indicates a genomic landscape in which the greatest diversity between species is represented in these SM clusters (28).
There has been considerable debate as to the role that gene clustering plays in the secondary metabolism of fungi. Such gene arrangement must be advantageous to the fungus; if natural selection did not favor clustering, one would assume that processes such as gene translocation and unequal crossing over would have caused dispersal over evolutionary history. Support for horizontal transfer from prokaryotes, in which genes are often arranged into operons, exists for the penicillin biosynthetic cluster (9) but not for other fungal clusters. A prokaryotic gene transfer hypothesis is weakened by the fact that fungal SM genes often contain introns and employ codon usage typical of other fungal genes. Another hypothesis holds that clustering provides a selective advantage to the cluster itself in that the arrangement makes propagation of the genes by means of horizontal transfer more successful (40). However, the importance of horizontal transfer of gene clusters among fungi has not been adequately explored to determine the merit of this hypothesis. This study focuses instead on the hypothesis that a common regulatory mechanism(s) underlies the SM cluster motif (25). Specifically, we have investigated whether clustering of SM biosynthetic genes allows coregulation through localized modification of chromatin structure.
SM clusters in Aspergillus species tend to be located near the telomeres of chromosomes (28), and a recent genome examination of the rice blast fungus Magnaporthe oryzae located at least two SM clusters within 40 kb of its telomeres (32). This locational bias may reflect in part an increased efficiency of epigenetic regulation at chromosomal subtelomeres—telomere-adjacent regions characterized by repeated DNA sequences (31). Though little is known about subtelomeric gene regulation events in aspergilli or other filamentous fungi, SM cluster regulation has been shown to be location dependent. Translocation of an A. parasiticus SM cluster gene to a chromosomal location outside of its native cluster can exempt it from coregulation with the rest of the cluster (14). The characterization of the protein LaeA also supports the case for chromatin-based regulation of SM clusters. LaeA acts as a global transcriptional regulator of SM clusters in several aspergilli and appears to be a protein methyltransferase with limited homology to histone methyltransferases (4). Importantly, LaeA also demonstrates a positional bias, as transfer of genes into or out of an SM cluster leads to gain or loss, respectively, of transcriptional regulation by LaeA (5). Chromatin regulation of gene expression is thought to be directed by modifications of histones, such as methylation and acetylation, that form the language of a combinatorial code. Histone modification patterns likely control the interaction of histones with transcriptional activators and repressors (23).
Arguably the most widely studied and best understood histone modification is acetylation. Histone acetylation states are dynamic and are controlled by the opposing actions of histone acetyltransferases and histone deacetylases (HDACs). As a general rule, hypoacetylation of histones tends to be associated with heterochromatin and gene silencing, while hyperacetylation is more commonly associated with euchromatin and gene activation (34, 39). Hypoacetylation of chromatin is also predominant in subtelomeric chromosomal regions (24). In the model fungus A. nidulans, the HDACs have been studied in particular detail (20, 38, 39). Therefore, this group of enzymes was chosen to study the effects of histone deacetylation on small-molecule production. We examined the effects of HDAC loss on the three best-characterized SM clusters in A. nidulans, the sterigmatocystin (ST) (a member of the carcinogenic and insecticidal aflatoxins) cluster (8), the penicillin (PN) (an antibiotic) cluster (7), and the terraquinone A (TR) (an antitumor agent) cluster (6). All three clusters are positively regulated by LaeA (3, 4). We created isogenic lines differing only in loss of one or more of the HDAC genes, hdaA, hosB, and hstA. The class 2 enzyme HdaA is responsible for the majority of HDAC activity in A. nidulans (38). HosB is an enzyme belonging to the HOS3-like subcategory of HDACs that is apparently unique to fungi (39), and HstA is an HDAC with homology to the NAD+-dependent sirtuin class. Sirtuin HDACs are known to be involved in the formation of heterochromatin in a broad range of species, including a number of fungi (10).
MATERIALS AND METHODS
Fungal strains.
Knockout procedures for hdaA, hosB, and laeA have been described previously (4, 38). hstA deletion mutants were generated by replacing the gene with the selection marker argB in the A. nidulans strain A89 (Table 1). PCR with primers Asir2kof and Asir2kor (Table 2) was used to amplify hstA and flanking sequence from the cosmid W23D09. The amplification product was ligated into a pGEM-T vector, and the coding sequence of hstA was eliminated with BamHI/NarI and replaced by a BamHI/ClaI-excised argB fragment. The resulting hstA deletion construct consisted of argB and ∼1,400 bp of the flanking region upstream and downstream of hstA. Transformation of A. nidulans was performed after elimination of the pGEM vector with ApaI and SpeI. The excised fragment was gel purified, and 7.5 μg of the DNA fragment was used for the transformation procedure. Putative deletion strains were verified by PCR and Southern blot analysis.
TABLE 1.
Genotypes of A. nidulans strains used in this study
| Strain | Genotype |
|---|---|
| Strains used in experimentsa | |
| RDIT 2.3 | veA1 |
| RJW 61.14 | ΔstcE::argB veA1 wA3 |
| RJW 61.1 | ΔlaeA::metG veA1 wA3 |
| RJW 61.9 | ΔlaeA::metG ΔstcE::argB veA1 wA3 |
| RJW 60.3 | ΔhdaA::pyrG veA1 wA3 |
| RJW 60.7 | ΔhdaA::pyrG ΔstcE::argB veA1 wA3 |
| RJW 60.1 | ΔhdaA::pyrG ΔlaeA::metG veA1 wA3 |
| RJW 60.4 | ΔhdaA::pyrG ΔlaeA::metG ΔstcE::argB veA1 wA3 |
| RJW 61.10 | ΔhstA::argB veA1 wA3 |
| RJW 61.5 | ΔhstA::argB ΔstcE::argB veA1 wA3 |
| RJW 61.12 | ΔhstA::argB ΔlaeA::metG veA1 wA3 |
| RJW 61.4 | ΔhstA::argB ΔlaeA::metG ΔstcE::argB veA1 wA3 |
| RJW 62.4 | ΔhosB::argB veA1 wA3 |
| RJW 62.2 | ΔhosB::argB ΔstcE::argB veA1 wA3 |
| RJW 62.8 | ΔhosB::argB ΔlaeA::metG veA1 wA3 |
| RJW 62.1 | ΔhosB::argB ΔlaeA::metG ΔstcE::argB veA1 wA3 |
| REKS 9.22 | ΔhdaA::pyrG ΔhstA::argB ΔhosB::argB ΔstcE::argB veA1 wA3 |
| REKS 9.21 | ΔhdaA::pyrG ΔhstA::argB ΔhosB::argB ΔlaeA::metG ΔstcE::argB veA1 wA3 |
| TJW 65.7 | ΔtdiB::pyrG pyroA4 veA1 |
| Strains for sexual crossesb | |
| A89 | biA1 argB2 veA1 |
| H4 | ΔhdaA::pyrG riboA1 chA1 yA2 veA1 pyrG89 |
| A3 | ΔhstA::argB biA1 veA1 argB2 |
| B6 | ΔhosB::argB biA1 veA1 argB2 |
| RJW 34.1 | ΔlaeA::metG ΔstcE::argB trpC801 pyrG89 veA1 wA3 |
| RDIT 30.12 | trpC801 argB2 metG1 veA1 |
| RDIT 55.12 | trpC801 argB2 metG1 pyrG89 pyroA4 veA1 |
| REKS 1.1 | ΔhdaA::pyrG argB2 pyroA4 veA1 |
Strains were used directly in experiments described in this paper.
Strains were used only for sexual crosses or transformation.
TABLE 2.
PCR primer sets used in hstA knockout and to generate probes for Northern blot assays
| Gene | Primer | Sequence |
|---|---|---|
| hstA | Asir2kof | 5′ACGAGAATATATCTCCCG3′ |
| Asir2kor | 5′CAACGCAAAGCATATATCG3′ | |
| Actin | ActF | 5′CTCTCCCCTTCTCTCCTCCACTT3′ |
| ActR | 5′CCGCACTCATGGTACTCCTGCTT3′ | |
| AN2647.3 | pen5F3 | 5′TTCTTGTGACGTAGGAATTGGCC3′ |
| pen5R3 | 5′TGATCTGTGAGCCTGGCCTCC3′ | |
| AN7830.3 | upSTF | 5′GAAGAGTTCTTGAATGAATTTGACG3′ |
| upSTR | 5′GCAGCGTAGGCATTTGCCCTC3′ | |
| penDE | penDEF | 5′ACGAATCCGGTTGGCATCGGC3′ |
| penDER | 5′TGAGCTCTGTGACCTGCTGGC3′ | |
| tdiA | tdiAF | 5′TCTGCTGCATCAGGCAGAGGC3′ |
| tdiAR | 5′TATTGATGCGGTGGATGATAGCG3′ | |
| tdiB | NAIf1 | 5′AGCACTCCTTCCTCCCTCGTG3′ |
| NAIr1 | 5′TCCTATACTTGCCACTCAGCCC3′ |
Table 1 lists all of the A. nidulans strains used for this study. All strains were maintained as glycerol stocks. Some strains are not discussed in the text but were used for sexual crosses to obtain the strains of interest. Sexual crosses of A. nidulans strains were conducted according to standard methods (30). Strain genotypes were identified by PCR amplification of the correct allele, followed by confirmation of the allele by Southern blot analysis according to standard procedures (36). Strains of Alternaria alternata and Penicillium expansum were wild-type strains provided by the laboratory of Craig Grau at the Department of Plant Pathology, University of Wisconsin—Madison.
SM analysis.
Published procedures were used to extract and analyze ST; its precursor; norsolorinic acid (NOR) (regulated identically to ST and derived from the same cluster) (12); PN; and TR (3, 4). Prior to ST and NOR extractions, all A. nidulans strains were point inoculated onto solid glucose minimal medium (GMM) in 10-cm-diameter petri dishes and incubated for 72 h at 37°C (37). ST extractions were also performed at 48 h, yielding similar results (data not shown). Prior to SM extraction, A. alternata and P. expansum strains were point inoculated onto solid GMM in 10-cm-diameter petri dishes and incubated for 72 h at 37°C on solid potato dextrose agar (Difco Laboratories, Sparks, MD). Solid-medium plates for all fungal strains were point inoculated with approximately 104 spores in 10 μl of water. For experiments involving H2O2, the chemical was added to media following autoclaving, after the media had cooled to 50°C; 30% (wt/wt) H2O2 in water was added to molten GMM agar to produce final concentrations of 0, 1, 2, or 3 mM. Dilutions of H2O2 were made with water so that an equal volume was added to the media to produce each of these concentrations. Prior to the PN assay, strains were inoculated into 50 ml liquid GMM at a spore concentration of approximately 106 spores/ml and incubated for 72 h at 37°C with shaking at 280 rpm. For experiments involving trichostatin A (TSA) (Invitrogen, San Diego, CA), the TSA was suspended in 100% ethanol and added to molten medium (GMM or potato dextrose agar) after it cooled to 50°C. TSA was dissolved in ethanol (0.45 mg/ml) and added to media to obtain a final concentration of 1 μM TSA. In control plates, an equal volume of ethanol was added. Prior to TR extraction, conidia of A. nidulans strains were inoculated into 50 ml liquid GMM to a concentration of approximately 106 conidia/ml and incubated for 72 h at 37°C with shaking at 280 rpm. For ST visualization, dried extracts from all culture types were resuspended in 100 μl of chloroform, and 5 μl was separated in a liquid phase consisting of toluene-ethyl acetate-acetic acid (8:1:1) on silica-coated thin-layer chromatography (TLC) plates. For TR visualization, a mixture of hexane-ethyl acetate (4:1) was used as the liquid phase for TLC. The ΔtdiB strain TJW 65.7 was used as a reference to determine the TR migration distance (3). Quantification of ST and TR from A. nidulans extracts and unknown compounds from A. alternata and P. expansum on TLC plates was accomplished using a CAMAG II densitometer, according to the manufacturer's instructions. A wavelength of 245 nm was used for all analyses. Quantification of PN from A. nidulans extracts was determined by measuring the diameter of bacterial clearing around each well containing extract (4). All experiments were performed in triplicate.
Nucleic acid analysis.
The extraction of DNA from fungi, restriction enzyme digestion, gel electrophoresis, blotting, hybridization, and probe preparation were performed by standard methods (36). Total RNAs were extracted from A. nidulans strains by use of Trizol reagent (Invitrogen) according to the manufacturer's instructions. Extractions were made from mycelia of cultures (106 spores/ml) grown in 50 ml liquid GMM at 37°C for 12, 24, 36, 48, or 72 h with shaking at 280 rpm. For experiments involving H2O2, the chemical was added to media following autoclaving, after the media had cooled to 37°C; 30% (wt/wt) H2O2 in water was added to 50 ml of liquid GMM to produce final concentrations of 0, 1, 2, or 3 mM. Dilutions of H2O2 were made with water so that an equal volume was added to the media to produce each of these concentrations. RNA blots were hybridized with a 0.7-kb SacII-KpnI fragment from the plasmid pRB7 containing the stcU coding region, a 1.3-kb EcoRV-XhoI fragment from plasmid pJW19 containing the aflR coding region, or a 1.1-kb EcoRI-HindIII fragment from the plasmid pUCHH(458) containing the ipnA coding region. PCR with gene-specific primers was used to generate probes for actin, AN2647.3, AN7830.3, penDE, tdiA, and tdiB. Primer sequences are shown in Table 2. All experiments were performed in duplicate or triplicate.
Statistical analysis.
For statistical analyses, a probability of type I error of less than 0.01 was considered statistically significant. Where only two treatments were compared, the significance of variation was determined using Student's t test. Where more than two treatments were compared, analysis of variance was used to determine the significance of overall variability among treatments, followed by a Neumann-Keuls test to compare individual pairs of treatments. The Microsoft Excel data analysis package was used to perform analysis of variance and t tests. Neumann-Keuls tests were performed by hand.
RESULTS
Location of clusters.
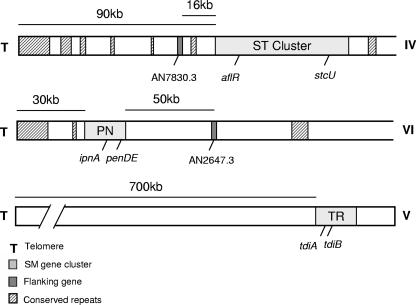
Several HDACs are known to regulate genes located in subtelomeric regions of other genomes (15, 19, 21, 22, 34, 41); thus, we were interested in identifying the locations of the three SM clusters in our study. Two of the three clusters are within 100 kb of the telomere: the ST cluster is 90 kb from the chromosome IV telomere, and the PN cluster is 30 kb from the chromosome VI telomere. In contrast, the TR cluster is 700 kb distal from the nearest telomere on chromosome V (Fig. 1). Both the ST and PN clusters functionally fulfill the definition of subtelomeric, as the intervening sequence between the clusters and telomere is characterized by repeated DNA found at the majority of A. nidulans chromosome ends. Such repeated sequences were not identified in the areas surrounding the TR cluster (Fig. 1). Subtelomere lengths vary considerably among eukaryotes; while Kluyveromyces lactis subtelomeres span only about 30 kb (17), those of humans may reach over 300 kb in length (33), and in trypanosomes, subtelomeric regions may make up the majority of the chromosome (13).
FIG. 1.
Chromosomal locations of ST, PN, and TR gene clusters and flanking genes. The Roman numerals to the right of the diagram indicate chromosome numbers. Distances are not to scale.
Production of SMs and expression of biosynthetic genes in the ΔhdaA mutant.
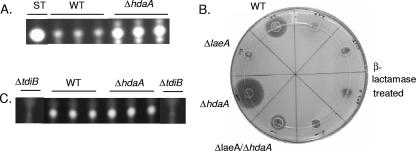
Prior experimentation had shown HdaA to exhibit most of the detectable HDAC activity in A. nidulans (38), and thus, our first studies examined the effect of loss of this allele on SM production. Production levels of ST; its precursor, NOR; PN; and TR were compared between the wild type and the ΔhdaA strain. The ΔhdaA mutant, which showed a wild-type growth rate, had increased production of subtelomeric metabolites (ST and PN) but unaltered TR levels (Fig. 2). As expected, NOR production paralleled ST production in the mutant (data not shown).
FIG. 2.
Production of ST, PN, and TR in the ΔhdaA mutant. (A) TLC plate showing ST extracted from wild-type (WT) and ΔhdaA strains. ST was extracted from 72-h cultures on solid media in triplicate. Similar results were obtained from 48-h cultures (data not shown). (B) Bacterial growth inhibition assay plate showing PN production by WT and ΔhdaA strains in laeA and ΔlaeA genetic backgrounds. The relative sizes of bacterial growth inhibition zones correspond to relative production of PN. The right half of the plate is a control treated with β-lactamase. PN was extracted from 72-h liquid shake cultures in triplicate and quantified using a bacterial growth inhibition assay. (C) TLC showing production of TR by WT and ΔhdaA strains. TR was extracted from 72-h cultures in liquid media in triplicate. The ΔtdiB strain, which does produce TR (3), was used as a reference to determine the TR migration distance.
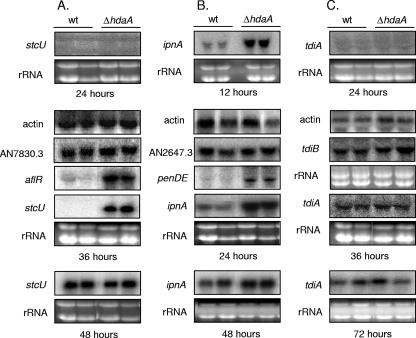
We used representative genes of the ST and PN clusters to examine whether the increased ST and PN production in the ΔhdaA strain correlated with mRNA production. For the ST cluster, these genes included aflR, which encodes a DNA-binding transcription factor required for expression of biosynthetic genes in the cluster (18, 43), and stcU (formerly verA), encoding a ketoreductase essential for ST production (26). aflR expression should correlate with expression of the entire cluster, while expression of stcU serves as an additional check, as this gene is known to be under the control of AflR and the two genes are located toward opposite ends of the cluster (Fig. 1). For the PN cluster, we examined two of the three genes in the cluster, ipnA and penDE (aatA), encoding an isopenicillin N synthetase and an isopenicillin N acyltransferase, respectively. Both genes are essential for PN production (7).
Transcription of the ST cluster is not readily observed until between 40 and 48 h of growth in the wild type (Fig. 3A), but stcU and aflR were strongly up-regulated in the ΔhdaA strain after only 36 h of growth, in contrast to no effect on an adjacent noncluster gene (AN7830.3) (Fig. 1 and 3A) and actin control (Fig. 3A). Neither the wild type nor the ΔhdaA strain showed transcription of ST genes at 24 h, and by 48 h, both strains showed approximately equal amounts of ST gene expression (Fig. 3A). Similar results were obtained for the PN cluster. Although some level of PN cluster expression was observed in the wild type at even the earliest time points examined, transcription of ipnA and penDE was considerably up-regulated in the ΔhdaA strain at 24 h (Fig. 3B), in contrast to an adjacent non-PN cluster gene (AN2647.3) (Fig. 1 and 3B) and actin control (Fig. 3B). ipnA transcription was also strongly increased at 12 h, but by 48 h, expression in the ΔhdaA strain was only slightly above wild type (Fig. 3B). Thus, in the cases of both the ST and PN clusters, HdaA appears to be involved in suppressing cluster expression during early stages of fungal development. As expected, expression of genes in the TR cluster, tdiA and tdiB, was not affected by hdaA loss (Fig. 3C). Thus, HdaA regulation specifically targets the two subtelomeric SM clusters.
FIG. 3.
Expression of ST, PN, and TR cluster genes in the ΔhdaA mutant. (A) Northern blots showing expression of the ST cluster genes stcU and aflR, as well as AN7830.3 (a gene approximately 16 kb telomere proximal from the ST cluster), and actin, by wild-type (wt) and ΔhdaA strains of A. nidulans. RNA was extracted after 24, 36, or 48 h of growth in liquid shake cultures in duplicate. (B) Northern blots showing expression of the PN cluster genes ipnA and penDE, as well as AN2647.3 (a gene approximately 50 kb telomere distal from the PN cluster) and actin, by WT and ΔhdaA strains of A. nidulans. RNA was extracted after 12, 24, or 48 h of growth in liquid shake cultures in duplicate. (C) Northern blots showing expression of the TR cluster genes tdiA and tdiB, as well as actin. RNA was extracted after 24, 36, or 72 h of growth in liquid shake cultures in duplicate.
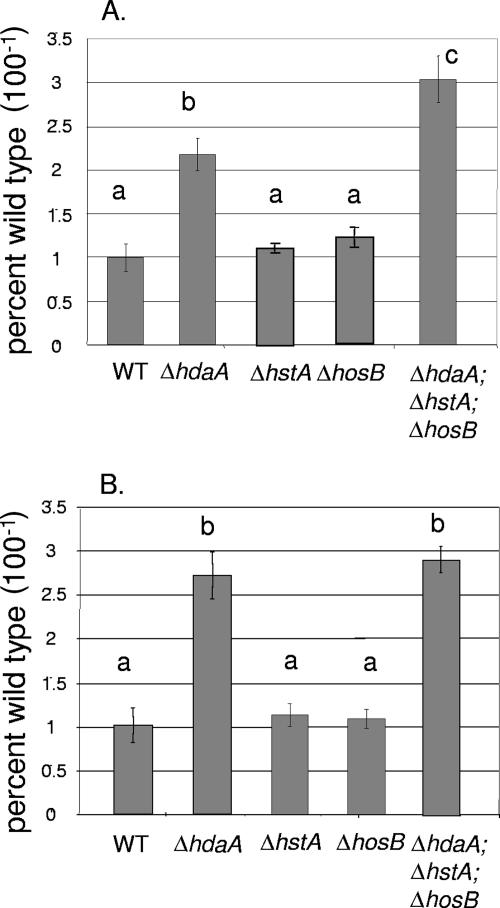
HosB and HstA contributions to SM regulation.
We were also interested in determining if other HDACs affected expression of the two HdaA-regulated clusters. NOR and PN production were examined and compared in all single HDAC mutants, as well as a triple mutant created by sexual cross. As shown in Fig. 4, only the ΔhdaA strain had a significant effect on metabolite production (an approximately 2-fold increase in NOR and 2.5-fold increase in PN) over the wild type. However, a mutant in which all three HDAC genes were nonfunctional showed an approximately threefold increase in production of NOR compared to the wild type (Fig. 4A), suggesting that the effects of the HDACs may be additive for some clusters.
FIG. 4.
Production of NOR and PN in the ΔhstA and ΔhosB mutants. (A) TLC plates were scanned for quantification of NOR by wild-type (WT), ΔhdaA, ΔhstA, ΔhosB, and triple HDAC mutant strains and depicted as a histogram. The growth conditions were the same as those in Fig. 2A. (B) Histogram showing relative production of PN by WT, ΔhdaA, ΔhstA, ΔhosB, and triple HDAC mutant strains. Growth and assay conditions were the same as those in Fig. 2B. For the histograms, WT production levels were assigned a value of 1, and all other production levels are presented relative to the WT. Different letters above the bars represent statistical differences at P < 0.01. The error bars represent ±1 standard deviation.
Oxidative stress and SM cluster expression.
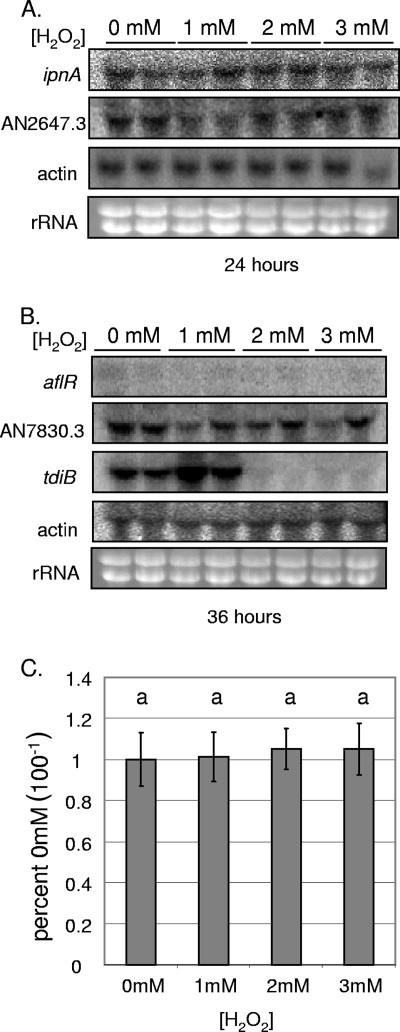
As the ΔhdaA mutant is known to have increased susceptibility to oxidative stress due to deficient production of the enzyme catalase (38), we considered the possibility that the observed effects of this mutation on SM cluster expression might be the result of increased oxidative stress. In order to determine if oxidative stress mimics the gene expression phenotype of the ΔhdaA strain, we examined expression of the ST, PN, and TR cluster genes in cultures of wild-type A. nidulans grown in liquid media containing concentrations of the oxygen radical-producing compound H2O2 ranging from 0 mM to 3 mM (Fig. 5) at times showing the greatest ΔhdaA effects (e.g., 36 h for the ST cluster and 24 h for the PN cluster) (Fig. 3). Figure 5 demonstrates that neither aflR nor ipnA was affected by H2O2 treatment, nor were flanking genes or actin. Concomitantly, H2O2. had no discernible effect on ST production (Fig. 5C). Interestingly, H2O2 did appear to affect expression of tdiB, for which low concentrations stimulated and high concentrations inhibited expression (Fig. 5B). As the presence of H2O2 does not produce effects resembling those of the ΔhdaA strain, we conclude that the effects on SM cluster expression observed in this mutant are not the result of oxidative stress.
FIG. 5.
Expression of ST, PN, and TR cluster genes in media containing H2O2. (A) Northern blots showing expression of the PN cluster gene ipnA, as well as actin and the PN cluster flanking gene AN2647.3, by wild-type A. nidulans grown for 24 h in medium containing, 0, 1, 2, or 3 mM H2O2. (B) Northern blots showing expression of the ST cluster gene aflR, the ST cluster flanking gene AN7830.3, the TR cluster gene tdiB, and actin by wild-type A. nidulans grown for 36 h in media with the same concentrations of H2O2 as in panel A. RNA was extracted from liquid shake cultures in duplicate. (C) Histogram depicting quantified TLC data for ST production by wild-type A. nidulans after 72 h of growth on solid media containing the aforementioned concentrations of H2O2. Treatments were performed in triplicate. Production levels at 0 mM H2O2 were assigned a value of 1, and all other production levels are presented relative to this. Different letters above the bars represent statistical differences at P < 0.01. The error bars represent ±1 standard deviation.
Effects of HDAC loss on the ΔlaeA phenotype.
Since loss of laeA (ΔlaeA) leads to a repression of global SM production in all aspergilli examined (4), we investigated whether LaeA might function by interfering with and/or activating heterochromatin formation, postulating that the ΔlaeA phenotype would be rescued by HDAC loss. Addition of ΔhdaA to a ΔlaeA background resulted in wild-type levels of NOR and PN production in the double mutant (Fig. 6); however, the increase in production was not equivalent to that of the ΔhdaA mutation alone. Thus, the loss of LaeA appears not to affect the function of HdaA, and vice versa. This suggests that, although HdaA and LaeA have opposing effects on ST and PN production, they operate through different mechanisms. This observation is supported by the fact that LaeA, unlike HdaA, positively regulates the telomere-distal TR cluster in addition to the two subtelomeric clusters (3, 4). Global regulation of both telomere-proximal and -distal SM clusters by LaeA is also seen in A. fumigatus (29).
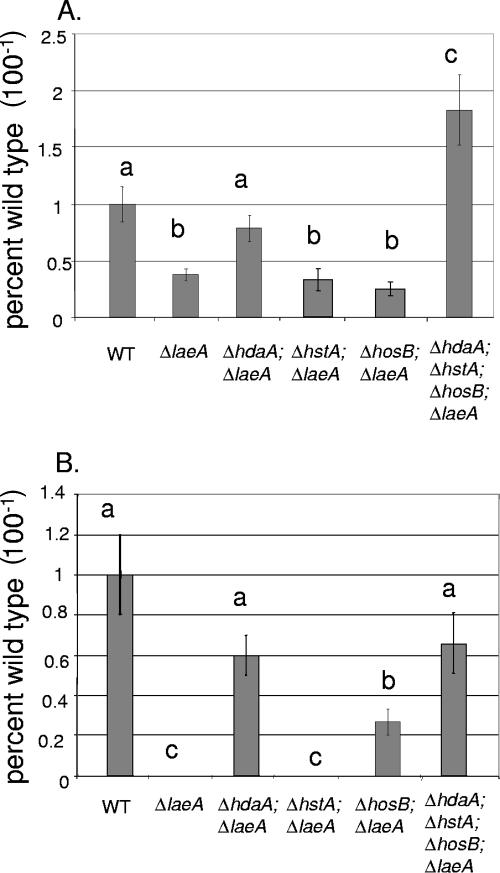
FIG. 6.
Production of NOR and PN in HDAC mutant strains with ΔlaeA genetic backgrounds. (A) Production of NOR by ΔlaeA mutant strains with additional individual ΔhdaA, ΔhstA, or ΔhosB mutations or with all three HDAC knockouts. NOR was extracted from 72-h cultures on solid media in triplicate. (B) Production of PN by ΔlaeA mutant strains with additional individual ΔhdaA, ΔhstA, or ΔhosB mutations or with all three HDAC knockouts. PN was extracted from 72-h liquid shake cultures in triplicate and quantified using a bacterial growth inhibition assay. For the histograms, wild-type (WT) production levels were assigned a value of 1, and all other production levels are presented relative to the WT. Different letters above the bars represent statistical differences at P < 0.01. The error bars represent ±1 standard deviation.
As the single ΔhstA and ΔhosB mutations did not affect production of NOR or PN (Fig. 4), we did not expect to see any effect on metabolite production when these alleles were placed in the ΔlaeA background. Unexpectedly, the ΔhosB ΔlaeA mutant did show increased production of PN over ΔlaeA alone, suggesting that this HDAC may play a role in regulating PN production under some circumstances. Combination of the three HDAC loss-of-function mutants with ΔlaeA increased NOR, but not PN, production over the ΔhdaA ΔlaeA double mutant, again arguing for an additive effect of all three HDACs on repression of the ST cluster.
Effects of HDAC inhibition on SM production in other fungal genera.
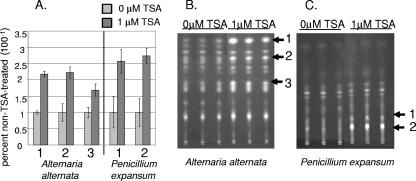
To determine whether HDAC regulation of SM clusters might extend to other species, we investigated fungi of two genera notable for their small-molecule arsenals. Representative isolates of the species A. alternata and P. expansum were treated with the class 1 and class 2 HDAC inhibitor TSA (35, 42). TSA treatment resulted in a statistically significant (P < 0.01) increase of numerous unidentified SMs in both species (Fig. 7). Thus, HDACs may function in the regulation of secondary metabolism among a broad range of fungal genera.
FIG. 7.
Effects of TSA on secondary metabolism of A. alternata and P. expansum. (A) Histogram of relative SM production levels in TSA-treated and untreated cultures. SM production levels in the absence of TSA were assigned a value of 1, and TSA-treated production levels are presented relative to untreated levels. The numbers on the x axis of the graph correspond to metabolites indicated on the TLC plates shown in panels B and C. The differences presented for individual compounds represent statistical differences at P < 0.01. The error bars represent ±1 standard deviation. SMs were extracted from 72-h cultures on solid media with or without 1 μM TSA in triplicate.
DISCUSSION
The data presented here constitute strong evidence for a role for HdaA in suppression of cluster-derived small-molecule production in A. nidulans and suggest that a role for HDAC in SM regulation may be conserved in other filamentous fungi. Elimination of HdaA, a major HDAC of A. nidulans, results in early and increased gene expression of two telomere-proximal small-molecule clusters and production of their corresponding metabolites. Transcriptional suppression by HdaA is precise, since neither actin nor the nearest expressed flanking genes to the ST and PN gene clusters were up-regulated in the ΔhdaA mutant. In this respect, HdaA presents cluster demarcation specificity similar, although in an opposing fashion, to that of LaeA (3, 6). However, in contrast to LaeA, where location of LaeA-regulated clusters ranges from telomere proximal to internal on chromosomal arms in both A. nidulans and A. fumigatus (3, 29), our current evidence supports a role for HdaA only in regulating telomere-proximal clusters.
This is congruent with findings in Saccharomyces cerevisiae, which, though devoid of SM clusters, does exhibit HDAC-dependent regulation of select subtelomeric genes. Hda1, the S. cerevisiae homologue of HdaA, demonstrates an overall targeting bias for telomere-proximal genes (34). This HDAC is known to be involved in silencing of the subtelomeric adhesin gene FLO11. Significantly, relocation of FLO11 to a telomere-distal region has been shown to release the gene from epigenetic silencing, suggesting that proximity to the telomere is important for such silencing to occur (21). Hda1 also is known to regulate clusters of metabolically unrelated subtelomeric genes, known as HAST (Hda1-affected subtelomeric) domains. These are comprised of a variety of genes that are activated in response to adverse environmental conditions, including genes involved in gluconeogenesis, fermentation, alternate carbon source utilization, and responses to various types of stress (34).
Epigenetic regulation of subtelomeric regions is also critical in the pathogenicity of several microbes. A prime example is the regulation of the subtelomeric var gene clusters of the malaria agent Plasmodium falciparum. HDAC-mediated regulation of these genes allows the protozoan to vary antigen display on the surfaces of infected host cells, thus evading the immune response (19). The subtelomeric EPA gene clusters of Candida glabrata, involved in biofilm formation and essential for pathogenicity, are regulated epigenetically by the NAD+-dependent HDAC Sir2p (15). These genes are turned on in response to the low-nicotinic acid environment of the host urinary tract, resulting in adhesion and subsequent infection (16). Thus, this system allows adhesin production only in the proper environment.
Although it is the sirtuin HDAC that is involved in C. glabrata subtelomeric gene regulation, under our conditions, we did not find a significant effect of hstA (or hosB) loss on NOR or PN production. However, the combination of these deletions with ΔhdaA did cause an increase in NOR production (Fig. 4), and the combination of ΔhosB with ΔlaeA also resulted in greater production of PN than in the ΔlaeA mutant alone (Fig. 6). It is interesting that ΔhstA and ΔhosB demonstrate a synergistic effect with ΔhdaA, but neither of the former HDAC mutants has a significant individual effect on SM production. Studies with yeast have shown that cooperative repression of chromosomal regions by multiple HDACs is common. While the S. cerevisiae HDACs Hda1, Rpd3 (a homolgue of the A. nidulans HDAC RpdA [20]), and Sir2 (a homologue of HstA) are each involved in the suppression of a unique set of genes, they also contribute to the silencing of numerous shared genes (1). Likewise, the Schizosaccharomyces pombe HDACs Clr3, Clr6, Sir2, and Hos2 (homologous to A. nidulans HdaA, RpdA, HstA, and HosA [20], respectively) repress both unique and shared genes (22, 41). The genes repressed by Clr3 and Sir2 demonstrate particularly strong overlap and also tend to overlap with genes regulated by the heterochromatin-associated protein Swi6 (41). Similar to the pattern we observed for NOR production in the triple HDAC mutant (Fig. 4), disruption of the genes encoding both S. pombe Clr3 and Clr6 results in upregulation of many genes to a degree higher than that resulting from individual mutation of either gene, and often to an extent greater than a merely additive effect. A large portion of these synergistically regulated genes were found to be subtelomeric (22). While the effects of hstA and hosB on ST/NOR and PN production were not as dramatic as those observed for hdaA, this is not to say that other natural products are not more strongly affected by mutation of these genes. More comprehensive studies are required to determine the extent to which these HDACs are involved in SM regulation. It should be noted that A. nidulans also possesses at least two additional HDACs, RpdA and HosA, that may well be important in this complex regulatory process.
Our data regarding the effects of the HDAC inhibitor TSA on the secondary metabolism of Fusarium and Penicillium provide evidence that HDAC-mediated regulation of small-molecule production may be a widespread phenomenon in filamentous fungi, which we speculate may have evolved as a tool to allow SM production under optimal environmental conditions. Pragmatically, as a variety of chemical HDAC inhibitors are readily available, treatment of fungi with such compounds could potentially provide a means of increasing production of beneficial metabolites and could also aid in the identification of novel natural products that may not have been previously detected due to low production levels under normal growth conditions.
The findings of our study support a role for epigenetic regulation of SM clusters. We hypothesize that epigenetic regulation of secondary metabolism is an efficient way for filamentous fungi to ensure that energetically costly molecules are synthesized only when production is likely to be advantageous. For example, HdaA-mediated repression of SM clusters occurs early in development (Fig. 3). It has long been observed that SM production is generally nil in the initial exponential growth phase and increases when nutrients are limited and growth is restricted (11). A global mechanism(s) to suppress expression of SM clusters during initial vegetative growth and yet allow SM production once sufficient biomass is established should yield competitive advantages for filamentous fungi and provide the fungus with an efficient means of responding to changing foraging and competitive pressures. Evidence suggests that at least two such mechanisms may be operating in the aspergilli: telomere-proximal SM cluster suppression by HdaA and a less spatially limited positive regulation by LaeA. Certainly, loss of laeA in A. fumigatus yields a less pathogenic organism with increased vegetative growth (2, 29).
Acknowledgments
Funding has been provided for this research and publication by the USDA Cooperative State Research, Education and Extension Service (CSREES) project WIS049621, NSF MCB-0236393, and NIH 1 R01 Al065728-01 to N.K., as well as the Austrian Science Foundation (P19750) and Tyrolean Science Foundation (0404/225) to S.G.
This article is dedicated to the memory of Ann Henry Keller.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Bernstein, B. E., J. K. Tong, and S. L. Schreiber. 2000. Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97:13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bok, J. W., S. A. Balajee, K. A. Marr, D. Andes, K. F. Nielsen, J. C. Frisvad, and N. P. Keller. 2005. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 4:1574-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bok, J. W., D. Hoffmeister, L. A. Maggio-Hall, R. Murillo, J. D. Glasner, and N. P. Keller. 2006. Genomic mining for Aspergillus natural products. Chem. Biol. 13:31-37. [DOI] [PubMed] [Google Scholar]
- 4.Bok, J. W., and N. P. Keller. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bok, J. W., D. Noordermeer, S. P. Kale, and N. P. Keller. 2006. Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol. Microbiol. 61:1636-1645. [DOI] [PubMed] [Google Scholar]
- 6.Bouhired, S., M. Weber, A. Kempf-Sontag, N. P. Keller, and D. Hoffmeister. 8 January 2007. Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator LaeA. Fungal Genet. Biol. doi: 10.1016/j.fgb.2006.12.010. [Epub ahead of print.] [DOI] [PubMed]
- 7.Brakhage, A. A. 1997. Molecular regulation of penicillin biosynthesis in Aspergillus (Emericella) nidulans. FEMS Microbiol. Lett. 148:1-10. [DOI] [PubMed] [Google Scholar]
- 8.Brown, D. W., J.-H. Yu, H. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five co-regulated transcripts define the sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buades, C., and A. Moya. 1996. Phylogenetic analysis of the isopenicillin-N-synthetase horizontal gene transfer. J. Mol. Evol. 42:537-542. [DOI] [PubMed] [Google Scholar]
- 10.Buck, S. W., C. M. Gallo, and J. S. Smith. 2004. Diversity in the Sir2 family of protein deacetylases. J. Leukoc. Biol. 75:939-950. [DOI] [PubMed] [Google Scholar]
- 11.Bu'Lock, J. D. 1961. Intermediary metabolism and antibiotic synthesis. Adv. Appl. Microbiol. 3:293-342. [DOI] [PubMed] [Google Scholar]
- 12.Butchko, R. A., T. H. Adams, and N. P. Keller. 1999. Aspergillus nidulans mutants defective in stc gene cluster regulation. Genetics 153:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callejas, S., V. Leech, C. Reitter, and S. Melville. 2006. Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length. Genome Res. 16:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiou, C. H., M. Miller, D. L. Wilson, F. Trail, and J. E. Linz. 2002. Chromosomal location plays a role in regulation of aflatoxin gene expression in Aspergillus parasiticus. Appl. Environ. Microbiol. 68:306-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Las Penas, A., S. J. Pan, I. Castano, J. Alder, R. Cregg, and B. P. Cormack. 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17:2245-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domergue, R., I. Castano, A. De Las Penas, M. Zupancic, V. Lockatell, J. R. Hebel, D. Johnson, and B. P. Cormack. 2005. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308:866-870. [DOI] [PubMed] [Google Scholar]
- 17.Fairhead, C., and B. Dujon. 2006. Structure of Kluyveromyces lactis subtelomeres: duplications and gene content. FEMS Yeast Res. 6:428-441. [DOI] [PubMed] [Google Scholar]
- 18.Fernandes, M., N. P. Keller, and T. H. Adams. 1998. Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol. 28:1355-1365. [DOI] [PubMed] [Google Scholar]
- 19.Freitas, L. H., Jr., R. Hernandez-Rivas, S. A. Ralph, D. Montiel-Condado, O. K. Ruvalcaba-Salazar, A. P. Rojas-Meza, L. Mancio-Silva, R. J. Leal-Silvestre, A. M. Gontijo, S. Shorte, and A. Scherf. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121:25-36. [DOI] [PubMed] [Google Scholar]
- 20.Graessle, S., M. Dangl, H. Haas, K. Mair, P. Trojer, E. M. Brandtner, J. D. Walton, P. Loidl, and G. Brosch. 2000. Characterization of two putative histone deacetylase genes from Aspergillus nidulans. Biochim. Biophys. Acta 1492:120-126. [DOI] [PubMed] [Google Scholar]
- 21.Halme, A., S. Bumgarner, C. Styles, and G. R. Fink. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116:405-415. [DOI] [PubMed] [Google Scholar]
- 22.Hansen, K. R., G. Burns, J. Mata, T. A. Volpe, R. A. Martienssen, J. Bahler, and G. Thon. 2005. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol. Cell. Biol. 25:590-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 24.Katan-Khaykovich, Y., and K. Struhl. 2005. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 24:2138-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 26.Keller, N. P., N. J. Kantz, and T. H. Adams. 1994. Aspergillus nidulans verA is required for production of the mycotoxin sterigmatocystin. Appl. Environ. Microbiol. 60:1444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller, N. P., G. Turner, and J. W. Bennett. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 3:937-947. [DOI] [PubMed] [Google Scholar]
- 28.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 29.Perrin, R., N. Federova, J.-W. Bok, J. Wortman, R. A. Cramer, Jr., H. Kim, W. Nierman, and N. Keller. 2007. Mapping the chromosomal landscape of co-regulated pathogenicity factors in Aspergillus fumigatus. PLoS Pathog. 3:e50.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. P. Macdonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 31.Pryde, F. E., H. C. Gorham, and E. J. Louis. 1997. Chromosome ends: all the same under their caps. Curr. Opin. Genet. Dev. 7:822-828. [DOI] [PubMed] [Google Scholar]
- 32.Rehmeyer, C., W. Li, M. Kusaba, Y. S. Kim, D. Brown, C. Staben, R. Dean, and M. Farman. 2006. Organization of chromosome ends in the rice blast fungus, Magnaporthe oryzae. Nucleic Acids Res. 34:4685-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riethman, H., A. Ambrosini, and S. Paul. 2005. Human subtelomere structure and variation. Chromosome Res. 13:505-515. [DOI] [PubMed] [Google Scholar]
- 34.Robyr, D., Y. Suka, I. Xenarios, S. K. Kurdistani, A. Wang, N. Suka, and M. Grunstein. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109:437-446. [DOI] [PubMed] [Google Scholar]
- 35.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93:14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tribus, M., J. Galehr, P. Trojer, G. Brosch, P. Loidl, F. Marx, H. Haas, and S. Graessle. 2005. HdaA, a major class 2 histone deacetylase of Aspergillus nidulans, affects growth under conditions of oxidative stress. Eukaryot. Cell. 4:1736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trojer, P., E. M. Brandtner, G. Brosch, P. Loidl, J. Galehr, R. Linzmaier, H. Haas, K. Mair, M. Tribus, and S. Graessle. 2003. Histone deacetylases in fungi: novel members, new facts. Nucleic Acids Res. 31:3971-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walton, J. D. 2000. Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: an hypothesis. Fungal Genet. Biol. 30:167-171. [DOI] [PubMed] [Google Scholar]
- 41.Wiren, M., R. A. Silverstein, I. Sinha, J. Walfridsson, H. M. Lee, P. Laurenson, L. Pillus, D. Robyr, M. Grunstein, and K. Ekwall. 2005. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 24:2906-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 43.Yu, J. H., R. A. Butchko, M. Fernandes, N. P. Keller, T. J. Leonard, and T. H. Adams. 1996. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet. 29:549-555. [DOI] [PubMed] [Google Scholar]