Abstract
The alb1 (pksP) gene has been reported as a virulence factor controlling the pigmentation and morphology of conidia in Aspergillus fumigatus. A recent report suggested that laeA regulates alb1 expression and conidial morphology but not pigmentation in the A. fumigatus strain AF293. laeA has also been reported to regulate the synthesis of secondary metabolites, such as gliotoxin. We compared the role of laeA in the regulation of conidial morphology and the expression of alb1 and gliP in strains B-5233 and AF293, which differ in colony morphology and nutritional requirements. Deletion of laeA did not affect conidial morphology or pigmentation in these strains, suggesting that laeA is not involved in alb1 regulation during conidial morphogenesis. Deletion of laeA, however, caused down-regulation of alb1 during mycelial growth in a liquid medium. Transcription of gliP, involved in the synthesis of gliotoxin, was drastically reduced in B-5233laeAΔ, and the gliotoxin level found in the culture filtrates was 20% of wild-type concentrations. While up-regulation of gliP in AF293 was comparable to that in B-5233, the relative mRNA level in AF293laeAΔ was about fourfold lower than that in B-5233laeAΔ. Strain B-5233laeAΔ caused slower onset of fatal infection in mice relative to that with B-5233. Histopathology of sections from lungs of infected mice corroborated the survival data. Culture filtrates from B-5233laeAΔ caused reduced death in thymoma cells and were less inhibitory to a respiratory burst of neutrophils than culture filtrates from B-5233. Our results suggest that while laeA is not involved in the regulation of alb1 function in conidial morphology, it regulates the synthesis of gliotoxin and the virulence of A. fumigatus.
Aspergillus fumigatus, a ubiquitous saprophyte, is the predominant cause of aspergillosis throughout the world. This species most often causes life-threatening invasive aspergillosis (IA) in individuals with dysfunctional phagocytes or with prolonged neutropenia resulting from immunosuppressive drug therapy (9, 19). In previous studies, we have dissected the molecular basis of conidial pigment formation and identified a cluster of six genes involved in the biosynthesis of dihydroxynaphthalene (DHN)-like melanin in A. fumigatus strain B-5233 (34-37). The alb1 (pksP) gene in this cluster encodes a polyketide synthase that catalyzes the first step of DHN-like melanin synthesis during conidial formation. Conidia of A. fumigatus are bluish green and coarsely echinulated due to the protrusions on their surfaces as revealed by scanning electron microscopy (34). Deletion of the alb1 gene from strain B-5233 resulted in two simultaneous changes in conidial morphology: albino instead of bluish green color and a smooth rather than echinulated surface. Complementation of the alb1Δ strain with the wild-type alb1 gene restored the bluish green pigment as well as the echinulation on the surface (17, 34), indicating that the alb1 gene is involved in conidial morphology as well as in conidial pigment synthesis. Furthermore, albino conidia were found to be more sensitive to hydrogen peroxide, were phagocytized more readily by neutrophils, and showed higher susceptibility to monocyte-mediated damage than wild-type conidia. More importantly, mice infected with albino conidia survived significantly longer than those infected with wild-type conidia (12, 17, 30, 34).
It has been reported recently that the function of alb1 is regulated by the LaeA protein (3). LaeA is a nuclear protein shown to control secondary metabolism in various aspergilli, including A. nidulans, A. terreus, and A. fumigatus. For instance, it has been demonstrated that the expression of laeA affects the synthesis of sterigmatocystin, penicillin, and lovastatin. Sequence analysis and experimental data suggested that the LaeA protein has methyltransferase activity and is predicted to function at the level of chromatin remodeling (3, 6, 7, 14). Upon deletion of laeA in strain AF293, the conidia became smooth, a result similar to that reported for the alb1Δ strain of B-5233 (34), without affecting the synthesis of bluish green pigment (3). This indicated that the pigmentation and the surface protrusions of conidia are controlled by separate factors. The laeA deletant strain of AF293, which we refer to as AF293laeAΔ, manifested an array of other features distinct from those of the wild type. These included impaired virulence associated with reduced levels of gliotoxin in the lungs of animals, higher susceptibilities of conidia and hyphae to host phagocytes, and the loss of dark pigment in mycelia submerged in an agar medium (3, 6).
Considering the involvement of laeA in the virulence of A. fumigatus and possibly in the regulation of alb1 function in conidial development, we investigated the role of laeA in the pathobiology of A. fumigatus strain B-5233, with special emphasis on conidial morphology. B-5233 is a highly virulent clinical strain isolated from a leukemic patient who succumbed to IA. This strain was used extensively for characterization of the antigenic properties useful for diagnosis of aspergillosis as well as for studies of host-Aspergillus interactions (10, 26, 27, 39) long before it was used to dissect the molecular control of conidial morphology and pigment synthesis (34-37). We deemed it important to study the role of laeA in this well-characterized strain because the genomic strain AF293, unlike other A. fumigatus strains, fails to grow robustly in defined minimal medium (MM) broth. Furthermore, a study by Bok and colleagues (3) suggests that conidial surface morphology and conidial pigmentation are controlled by different factors in AF293, while previous reports (17, 34) demonstrated that these characteristics are both controlled by the alb1 gene in B-5233. We deleted the laeA gene in strain B-5233, compared conidial morphology and transcriptional regulation of alb1 with those for the AF293laeAΔ strain, and observed that the laeA gene was not involved in the regulation of conidial morphology in these strains. However, laeA was involved in the expression of alb1 during the early stages of mycelial growth and showed a reduced amount of transcript at 48 h in both strains rather than a larger amount of transcript, as has been reported previously (3). laeA was also involved in the regulation of gliotoxin biosynthesis and the virulence of A. fumigatus. Our findings support the notion that laeA plays an important role in the regulation of gliotoxin production and the pathogenicity of A. fumigatus but not in the morphogenesis of conidia.
MATERIALS AND METHODS
Strains and media.
AF293 is a clinical isolate used for the sequencing of the A. fumigatus genome (22). B-5233 is also a clinical strain; it was isolated in a case of IA from a patient with leukemia (36). A. fumigatus strains were maintained on Aspergillus MM (28) or Sabouraud agar slants. Fungal strains were grown either in a liquid medium containing MM supplemented with 2 ml/liter of a vitamin mix (2 mg liter−1 p-aminobenzoic acid, 2 mg liter−1 niacin, 2 mg liter−1 pyridoxine HCl, 2 mg liter−1 riboflavin, 2 mg liter−1 thiamine HCl, 2 mg liter−1 choline HCl, 0.004 mg liter−1 d-biotin) or in RPMI 1640 medium. RPMI 1640 was used either alone or buffered with 25 mM HEPES, pH 7.2 (RPMI-HEPES). The medium was supplemented with 10 mM uracil and 10 mM uridine (UU) whenever strain AF293.1 (a pyrG1 mutant) was included in the experiments. Germination of conidia and the growth rate of mycelia were assayed by culturing the fungus in either Erlenmeyer flasks or 8-well chambered cover slides (Nalge Nunc International). For growth in flasks, 1 × 107 conidia were inoculated into 50 ml of broth (RPMI-HEPES or MM supplemented with vitamins) and incubated at 37°C on a shaker for 1 to 4 days. For growth on chambered cover slides, 3 × 105 conidia were inoculated into 200 μl of medium and incubated at 37°C under 5% CO2 for 24 to 40 h. The wells were then examined using a bright-field microscope. Strains AF293, AF293.1, and AF293laeAΔ (strain 54.2) were kindly provided by N. P. Keller, University of Wisconsin—Madison.
Transformation vectors. (i) Deletion vector.
Primers laeA1 (CGACTAGTCCTCGCTCCATCCCAATAGG) and laeA2 (CGTCTAGACGGAGTTGTTTCTTGAGCGG) were used to amplify a 777-bp fragment upstream (5′) of the laeA coding region, and primers laeA3 (CGAAGCTTACCGAGATTACCCTTGCATG) and laeA4 (CGCTCGAGCGACACACATATCATGACGG) were used to amplify a 990-bp fragment downstream (3′) of the coding region from the genomic DNA of strain B-5233. The deletion vector was constructed by cloning the 777-bp 5′ fragment, the 990-bp 3′ fragment, and a SacI/XbaI hygromycin resistance cassette from pAN7 into the SacI (blunted)/XhoI site of vector pDHt-SK (31). This deletion vector was used to transform strain B-5233 via Agrobacterium tumefaciens-mediated transformation (31).
(ii) Complementation vector.
Primers laeA10 (TGGAGCATAACCGAGTCTCC) and laeA11 (GAGGGATATACTGCCGTCCA) were used to amplify a 3.5-kb fragment that included 1.6 kb upstream and 0.73 kb downstream of the laeA coding region. The complementation vector was constructed by cloning the 3.5-kb fragment and the phleomycin resistance cassette into the HindIII/EcoRI-digested pDHt-SK vector. The complementation vector was used to transform strain B-5233laeAΔ via A. tumefaciens-mediated transformation.
SEM.
Conidia from 7-day-old cultures were fixed for 60 min with 4% (vol/vol) glutaraldehyde in phosphate-buffered saline (PBS) (pH 7.4) by adding the fixative to the cultures at a final concentration of 2%. One milliliter of this suspension was filtered through a 0.6-μm-pore-size polycarbonate membrane filter (Sterlitech, Kent, WA), and fixation was continued for an additional 60 min. The filters were washed three times with water for a period of 10 min each time, postfixed with 1% osmium tetroxide in water for 60 min, and washed three times as described above. The filters were serially dehydrated, submitted to critical point drying, mounted on aluminum stubs, and coated with a gold-palladium alloy. The samples were viewed under a Hitachi S-4700 field emission scanning electron microscope (SEM) (Hitachi High Technologies America, Inc., Schaumburg, IL) operated at an accelerating potential of 25 kV.
Quantitative real-time PCR.
One milliliter of a conidial suspension (107 conidia) was inoculated into 100 ml of RPMI-HEPES. The cultures were grown at 37°C under 5% CO2 with constant shaking for 24, 48, and 72 h. The RNA was isolated with Trizol (Invitrogen), purified with an RNeasy kit (QIAGEN), and treated with Turbo DNase (Ambion) according to the manufacturer's instructions. Typically 1 μg of the total RNA obtained was used for cDNA synthesis. The RNA was reverse transcribed using a high-capacity cDNA archive kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The first-strand cDNA was diluted 1:2, and 5 μl was added to 20 μl of the real-time PCR (RT-PCR) mixture, consisting of 12.5 μl of TaqMan universal PCR master mix (Roche Molecular Systems, CA), 2.25 μl (each) of 10 mM forward and reverse primers, 0.0625 μl of 10 mM probe, and 2.93 μl of water. The reaction was performed on the ABI Prism 7700 sequence detection system. Total RNA was used as a negative control. The glyceraldehyde-3-phosphate dehydrogenase gene, gpdA (Afu5g01970), was amplified as an endogenous control to standardize the amount of sample added to the reaction mixture. Primer and probe sequences for the following genes are given in parentheses: gpdA (probe, CCCCCATGTTCGTCATGGGTGTC; forward primer, TCTCCGCTCCTTCTGCTGAT; reverse primer, CGGAGGTGTAGGTGGTGTTGT), gliP (probe, CAATCCACCTTGGTCCTGGCCG; forward primer, CCTGAACGCCATGCACAAG; reverse primer, CCAGCCGGCGGTAGAAGT), and alb1 (probe, TGCGCAAACGCTTGTCGACCAC; forward primer, GCCATCGTCTCTCTACGCTGAT; reverse primer, CTGGTACTCTGGTTTGTATTTTGTGATC). The difference in the threshold cycle between gpdA in the wild-type and laeAΔ strains was less than 1 cycle (data not shown), indicating that gpdA expression was not affected by deletion of laeA and therefore that gpdA was suitable as an endogenous control.
Culture filtrate.
The culture filtrates used in the EL4 thymoma cell, mouse embryonic fibroblast (MEF), and gliotoxin quantification assays were prepared by inoculating 107 conidia into 100 ml of RPMI 1640 and incubating at 37°C under 5% CO2 for 48 h. Culture filtrates were then collected by filtering the culture through a sterile BD Falcon 40-μm-pore-size cell strainer (BD Biosciences, Erembodegem, Belgium). Culture filtrates for neutrophil chemiluminescence assays were prepared by inoculating 3 × 108 conidia into 100 ml of RPMI-HEPES and incubating at 37°C under 5% CO2 for 72 h before the culture supernatant was obtained.
Gliotoxin quantitation.
Aspergillus culture filtrates (100 ml), obtained as described above, were extracted with 100 ml of chloroform, and the organic phase was recovered, dried, and resuspended in 5 ml of chloroform. High-performance liquid chromatography (HPLC) analysis was performed as described elsewhere (2). Briefly, 100 μl of chloroform extract was injected in a reverse-phase C18 column (Waters) with a mobile phase (1 ml min−1) of methanol-water (70:30). Gliotoxin was quantified using a standard curve of the pure compound (Sigma, St. Louis, MO).
Neutrophil respiratory burst assay.
All human blood samples used in this study were collected after informed consent from healthy subjects (National Institutes of Health protocol 99-CC-0168). Blood was anticoagulated using acid citrate dextrose, and neutrophils were purified as described previously (42). The preparations were 95% pure. Twenty five microliters of RPMI-HEPES alone or containing 5 μl of a culture filtrate (5% of the final assay volume) was added to the wells of a white polypropylene 96-well plate (Whatman UNIPLATE). Twenty five microliters of 106 neutrophils ml−1 in RPMI-HEPES was added to each well and incubated at 37°C under 5% CO2. After 30 min, cells were quickly mixed with prewarmed 2× phorbol 12-myristate 13-acetate (PMA) diluted in a Diogenes chemiluminescent enhancer (National Diagnostics, Atlanta, GA), and relative light units (RLU) were measured for 0.5 s per well every 90 s in an Anthos Zenyth 3100 luminometer at 37°C with intermittent shaking. Raw data are presented kinetically as “sum RLU” (equaling the sum of all the measurements of RLU over the first 60 min) to facilitate comparison of different PMA doses and filtrate conditions.
Cell death.
EL4 cells (2 × 105) in 1 ml of minimum essential medium supplemented with 5% fetal calf serum were incubated for 16 h with 2.5, 5, 10, or 20% (vol/vol) culture filtrate. After the cells were washed with PBS, phosphatidylserine translocation and membrane integrity were monitored using the Annexin V (AV) detection kit (BD PharMingen, San Diego, CA) and propidium iodide (PI) staining as described previously (23). Cells positive for AV and PI and cells positive for AV but negative for PI were scored, and the sum of both is shown as a percentage of the total population. In every experiment, 104 cells were counted, and specific cell death was calculated by the following formula: [% (AV+ PI+ + AV+ PI− cells)] = [% (AV+ PI+ + AV+ PI− cells) in treated cells] − [% (AV+ PI+ + AV+ PI− cells) in the untreated control]. The percentage of the sum of AV+ PI+ and AV+ PI− cells in the control was always lower than 15%. Caspase-3 activation was monitored using a fluorescein isothiocyanate-labeled monoclonal antibody against the active form of the enzyme by fluorescence-activated cell sorting as described elsewhere (23).
Cell detachment.
MEFs transformed with simian virus 40 (24, 40) were cultured overnight in minimal essential medium supplemented with 10% fetal calf serum and 2-mercaptoethanol (10−5 M) at 37°C under 7% CO2. The medium was replaced by various dilutions of Aspergillus culture filtrate (2.5, 5, and 10%), and the MEFs were incubated for an additional 4 h before the cells were observed by bright-field microscopy. Photographs were taken using an Axiovert 10 microscope (Zeiss) with Axiovision software.
Virulence.
129/Sv mice were immunosuppressed by subcutaneous injection of 2 mg of hydrocortisone acetate in 100 μl of PBS-0.1% Tween 20 on days −4, −2, 0, 2, and 4. On day zero, mice were inoculated intranasally with 5 × 106 conidia in 20 μl of PBS-0.01% Tween 20. Twelve mice per treatment were used. Morbidity and mortality were monitored daily for 21 days, and subsequently the surviving mice were sacrificed. Control mice, immunosuppressed with hydrocortisone and inoculated intranasally with 20 μl of PBS-0.01% Tween 20, survived during the observation period without any sign of infection. Histopathological sections of lungs were prepared from mice that had been infected with B-5233, B-5233laeAΔ, or B-5233laeAR and sacrificed at 72 or 96 h after infection. The tissue sections were fixed, paraffin embedded, sectioned, and stained with hematoxylin and eosin or Gomori's methenamine silver.
RESULTS
Growth comparison between B-5233 and AF293.
Strain B-5233 grew robustly in MM broth with or without the vitamin mix and formed fungal balls (1 to 2 mm in diameter) within 24 h, whereas growth of strain AF293 in MM broth was not visible until after 4 days. Addition of the vitamin mix to MM broth allowed strain AF293 to form mycelia that were barely visible to the naked eye after 72 h. Since AF293 eventually formed mycelia in all the vitamin dropout media, none of the vitamins tested appeared to be essential for the growth of strain AF293. The assays on chambered slides showed that conidia of both B-5233 and AF293 germinated and formed a mycelial layer to an equal degree in the rich medium, RPMI-HEPES (Fig. 1A and B). The rates of germination and growth for the two strains were clearly distinguishable, however, when they were grown in MM broth with the vitamin mix. While B-5233 conidia germinated and formed a layer of mycelium (Fig. 1C), more than 90% of AF293 conidia remained ungerminated by 40 h (Fig. 1D). AF293 also showed slower growth on a solid agar medium containing MM alone or supplemented with the vitamin mix (Fig. 1E and data not shown). The severe growth retardation of AF293 in MM broth and not in a rich medium suggests that the strain is nutritionally deficient compared to B-5233. Furthermore, RPMI 1640 is better suited for growth of the genomic sequencing strain of A. fumigatus than the widely used Aspergillus MM with vitamins.
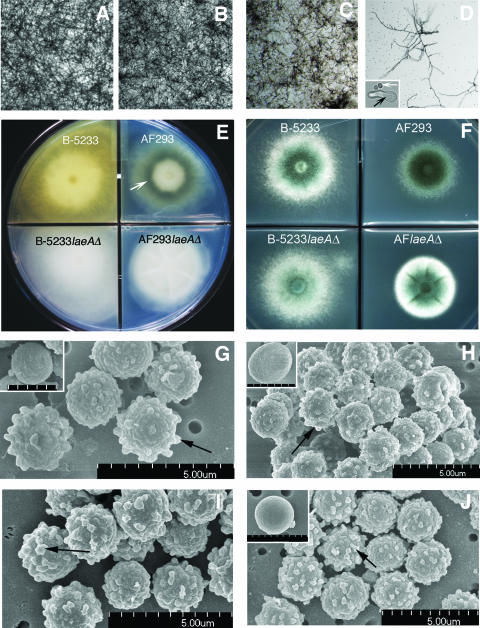
FIG. 1.
Phenotype of A. fumigatus strains. (A to D) Two different isolates from A. fumigatus, strains B-5233 and AF293, were grown in liquid media (chambered slide) at 37°C for 40 h. (A and B) B-5233 and AF293, respectively, grown on RPMI-HEPES. (C and D) B-5233 and AF293, respectively, grown on MM plus a vitamin mix. Panel D shows a few mycelia with branched hyphae. The majority of conidia remain ungerminated (small dots). (Inset) Nongerminated (white arrow) and germinated (arrow in black) conidia. Original magnifications, ×25 for panels A to D and ×1,000 for the inset in panel D. (E and F) AF293 and B-5233, and their respective laeA deletants, B-5233laeAΔ and AF293laeAΔ, grown on MM agar. (E) Growth at 25°C for 5 days showed a yellow compound secreted by B-5233 but not by B-5233laeAΔ, AF293, or AF293laeAΔ. Submerged mycelia of AF293 show dark pigmentation (white arrow) manifested in the colony reverse. (F) Strains were grown at 37°C for 2 days. (G to J) SEM of conidia from B-5233 (G), B-5233laeAΔ (I), AF293 (H), and AF293laeAΔ (J). Arrows indicate protrusions on the conidial surface. Scale bars for the insets in panels G, H, and J, 2.5, 2.8, and 3 μm, respectively.
Deletion of laeA and regulation of alb1 expression.
We and others have previously observed that alb1 is a virulence factor, that its expression is conidial stage specific, and that it catalyzes the synthesis of DHN-like melanin conidial color (17, 34). In addition to color, the alb1 gene also controls the surface morphology of conidia. Since laeA has been reported to control conidial morphology but not conidial color in AF293, we deleted laeA in order to investigate its regulatory role in the expression of alb1 in strain B-5233. laeA was deleted by replacing the coding region of the gene with a hygromycin resistance cassette. Southern blot hybridization analysis identified a transformant that had undergone homologous gene replacement, resulting in the deletion of laeA (data not shown). The deletant strain, designated B-5233laeAΔ, was then complemented with the wild-type laeA gene, and this reconstituted strain, which harbored a single copy of the gene, was named B-5233laeAR. A consistent phenotype of the laeA deletant that distinguished it from wild-type strain B-5233 was the absence of a yellow compound secreted in MM agar. Due to the absence of this yellow compound, the colony reverse of strain B-5233laeAΔ remained white (Fig. 1E, lower left panel), while those of strains B-5233 (Fig. 1E, upper left panel) and B-5233laeAR (data not shown) became yellow after 5 days of growth at 25°C. Strain AF293 showed a colony reverse with darkly pigmented mycelia and without secretion of the yellow compound (Fig. 1E, upper right panel). Unlike that of AF293, the colony reverse of AF293laeAΔ was white due to the loss of dark mycelial pigment (Fig. 1E, lower right panel). Other than the absence of the yellow exudates, deletion of laeA in B-5233 caused no significant phenotypic alteration in the size and morphology of the colony or in conidial pigmentation except for a slight delay in sporulation in cultures grown on MM agar (Fig. 1F). Furthermore, microscopic observation of conidia incubated in chambered cover slides for 8 h in RPMI-HEPES at 37°C showed comparable germination and hyphal emergence for B-5233 and B-5233laeAΔ (data not shown). Although the colony morphologies of B-5233 and AF293 were different on solid medium, the morphologies of their conidia examined under a compound microscope as well as by SEM were indistinguishable (Fig. 1G and H). Conidia of strains B-5233laeAΔ and AF293laeAΔ were morphologically identical to those of their parental strains, including the degree of protrusions (Fig. 1I and J). A more extensive search revealed rare smooth conidia devoid of protrusions for both the wild-type and the laeAΔ strains at similar frequencies (less than 0.1%), indicating that the surface morphology of conidia is unrelated to the laeA gene (Fig. 1G, H, and J insets).
Using Northern blot hybridization, Bok and collaborators (3) monitored the expression of alb1 in cultures grown in liquid MM for 24, 48, and 72 h and observed that the expression of alb1 at 48 h was significantly higher in strain AF293laeAΔ than in strain AF293. To assess the expression levels of alb1 in strain B-5233, we used MM as the growth medium and included strains AF293.1 and AF293laeAΔ as controls. AF293.1 was chosen because this strain was used as the background for the deletion of laeA (3). Strains AF293.1 and AF293laeAΔ grew extremely poorly in MM plus UU. Even when the medium was supplemented with a vitamin mix, strains with an AF293 background failed to attain growth comparable to that of B-5233 and B-5233laeAΔ within 48 h. Since it is essential to use a medium that can yield similar growth for the comparison of gene expression, we used the relatively rich medium RPMI 1640, which allowed comparable growth rates for all the strains (Fig. 1A and B and data not shown). The level of alb1 expression in each strain was analyzed by RT-PCR. At 48 h, strain B-5233laeAΔ exhibited a down-regulation of alb1 compared to levels in B-5233 and B-5233laeAR (Fig. 2A). A similar reduction in transcription, instead of an increase, was observed for strain AF293laeAΔ compared to AF293.1 (Fig. 2A). At 24 and 72 h, similar patterns of down-regulation were observed in the laeA deletant strains of B-5233 and AF293 (data not shown). These results indicate that although laeA does not interfere in the function of alb1 in conidial morphology, it affects the expression of alb1 during mycelial growth in a liquid medium.
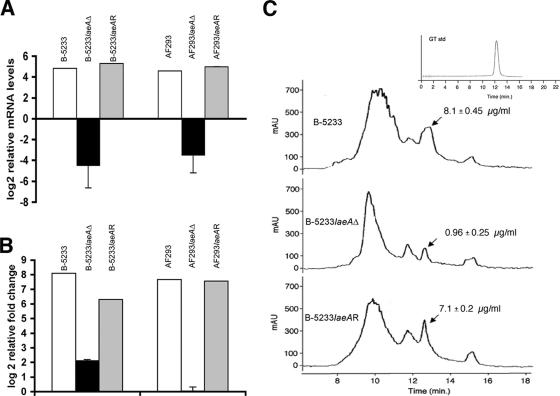
FIG. 2.
Transcriptional levels of alb1 and gliP and gliotoxin quantitation on culture filtrates. (A and B) Quantitative RT-PCR with RNAs isolated from cultures grown for 48 h in RPMI-HEPES. Shown are relative mRNA levels of alb1 (A) and gliP (B). Open bars, wild-type strains; solid bars, gene deletion strains; shaded bars, reconstituted strains. The mRNA levels were normalized to that of gpdA. The assay was repeated twice with similar results. (C) HPLC analysis of culture filtrates from strains B-5233, B-5233laeAΔ, and B-5233laeAR. Gliotoxin concentrations are means ± standard errors of the means from three independent experiments. Arrows indicate gliotoxin peaks as determined by comparison with a gliotoxin standard (inset in upper right corner). mAU, milli-absorbance units.
Deletion of laeA affects transcription of the gliP gene and gliotoxin synthesis.
We also monitored the transcriptional levels of the gliP gene, which is involved in the synthesis of gliotoxin, a secondary metabolite with immunosuppressive properties produced by A. fumigatus. The RT-PCR results from 48-h cultures showed that the expression level of gliP was significantly lower for B-5233laeAΔ than for B-5233 and B-5233laeAR. An even more accentuated difference was observed for strain AF293 (Fig. 2B). The RT-PCR results from the 24- and 72-h cultures showed the same pattern as that observed at 48 h (data not shown). Since the expression of gliP was diminished in the laeA deletants of both strains B-5233 and AF293, we investigated the effect of laeA on gliotoxin production using only strains B-5233, B-5233laeAΔ, and B-5233laeAR. Culture filtrates of the three strains were subjected to HPLC analysis to detect the secreted gliotoxin (Fig. 2C). The analysis showed that strain B-5233laeAΔ produced approximately 80% less gliotoxin than strains B-5233 and B-5233laeAR.
Effects of culture filtrates on the respiratory burst of neutrophils.
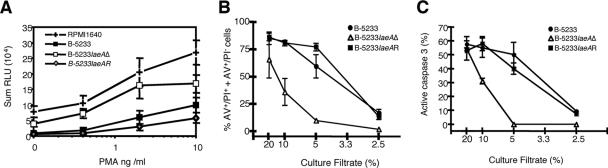
Since deletion of laeA caused a reduction in the synthesis of gliotoxin, the toxin known to inhibit the respiratory burst of neutrophils (38, 41), we analyzed the effects of the culture filtrates from B-5233, B-5233laeAΔ, and B-5233laeAR on superoxide anion production by human neutrophils. In the absence of culture filtrates, PMA stimulated neutrophil chemiluminescence in a dose-dependent manner (Fig. 3A), whereas preincubation of neutrophils with 5% of the filtrate from either B-5233 or B-5233laeAR significantly decreased the chemiluminescence but not the viability of neutrophils (Fig. 3A and data not shown). In contrast, the culture filtrate from strain B-5233laeAΔ had only a minor effect relative to results for the RPMI control. To exclude the possibility that the inhibitory activity present in B-5233 was due to a substance that interfered with the detection of chemiluminescence, we tested the effects of B-5233 and B-5233laeAΔ culture filtrates on chemiluminescence produced by xanthine oxidase/hypoxanthine, a cell-free superoxide-generating enzyme and substrate. Culture filtrates of the two strains had equivalent but negligible effects on light production (data not shown), suggesting that the inhibitory effect of the B-5233 culture filtrate was due to direct inhibition of the respiratory burst.
FIG. 3.
Effects of Aspergillus culture filtrates on neutrophil chemiluminescence and death of EL4 thymoma cells. (A) Neutrophils were incubated for 30 min with culture filtrates from strain B-5233, B-5233laeAΔ, or B-5233laeAR; PMA was then added to induce the oxidative burst, and chemiluminescence was measured. Data represent the means of the sums of RLU (n = 4 to 6 donors). Error bars, standard errors of the means. (B and C) Cell death was monitored using phosphatidylserine translocation, membrane integrity, and caspase-3 activation. EL4 cells were incubated for 16 h in 2.5, 5, 10, or 20% culture filtrates from B-5233, B-5233laeAΔ, or B-5233laeAR. (B) Phosphatidylserine translocation and membrane integrity were monitored with AV and PI, respectively. (C) Caspase-3 activity was used as a marker for apoptosis. Results for panels B and C are means ± standard errors of the means from three and two independent experiments, respectively.
Effects of culture filtrates on EL4 thymoma cells and MEFs.
Culture filtrates from A. fumigatus are known to cause damage to various types of mammalian cells (1, 13, 21). Therefore, we compared the effects of culture filtrates from strains B-5233, B-5233laeAΔ, and B-5233laeAR on proapoptotic processes in EL4 thymoma cells as well as on MEF adherence. EL4 thymoma cells were incubated with 2.5, 5, 10, or 20% culture filtrates before cell death was monitored. The results showed that the culture filtrates from all three strains caused cell death in a dose-dependent manner (Fig. 3B). At a culture filtrate concentration of 20%, the level of cell death caused by the B-5233laeAΔ filtrate was only slightly lower than that caused by the B-5233 culture filtrate. However, at intermediate concentrations, 10 and 5%, levels of cell death caused by the B-5233laeAΔ filtrate were six- and twofold lower, respectively, than those caused by the B-5233 filtrate. At the lowest concentration, 2.5%, the filtrate from B-5233laeAΔ had almost no effect (less than 2% cell death), whereas the filtrate from B-5233 was still able to cause death for approximately 15% of the cells. The number of cells showing active caspase-3 in the populations treated with 5 or 10% B-5233laeAΔ culture filtrates was at least 40% lower than that in the population treated with the filtrates of B-5233 (Fig. 3C). The culture filtrates of B-5233laeAR and B-5233 showed similar results, indicating that reconstitution of the laeA gene restored the wild-type phenotype. These results suggest that deletion of laeA reduces the ability of B-5233 to induce proapoptotic processes and cause mammalian cell death in vitro.
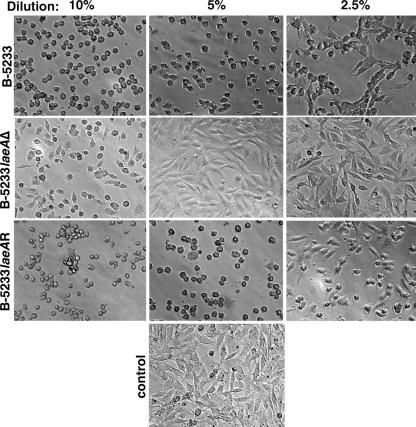
When MEFs were grown in RPMI, they formed a monolayer of spindle-shaped cells attached to the surface of the culture plate (Fig. 4, control). However, addition of as little as 2.5% of the culture filtrate from either B-5233 or B-5233laeAR caused some of the MEFs to display a rounded morphology and to detach from the plate. With 5% or a higher percentage of culture filtrates, a majority of the cells displayed the rounded morphology and detached from the surface. In contrast, no such morphological change or detachment of MEFs occurred with culture filtrates of B-5233laeAΔ until the concentration was elevated to 10%, suggesting that deletion of laeA affected the ability to cause MEF detachment.
FIG. 4.
A. fumigatus culture filtrates cause MEF detachment. MEFs were incubated for 4 h with different concentrations of the culture filtrates (2.5, 5, and 10%). The majority of the cells incubated in 5% culture filtrates from B-5233 and B-5233laeAR are rounded and detached from the culture plate surface, whereas cells incubated with RPMI only (control) are elongated and attached to the surface. Original magnification, ×100.
Virulence.
The virulence of strains B-5233, B-5233laeAΔ, and B-5233laeAR for 129/Sv mice immunosuppressed with hydrocortisone (33) was tested. We chose to use the mouse strain 129/Sv and corticosteroid immunosuppression because the data obtained with this model were more reproducible than those with C57BL/6 or BALB/c mice immunosuppressed with corticosteroids either alone or in combination with cytotoxic agents such as cyclophosphamide (J. Pardo and M. M. Simon, unpublished data). Mice infected with strain B-5233laeAΔ survived significantly longer (P = 0.0076 by the Kaplan-Meier survival curve and log rank test) than those infected with strain B-5233 or B-5233laeAR (Fig. 5).
FIG. 5.
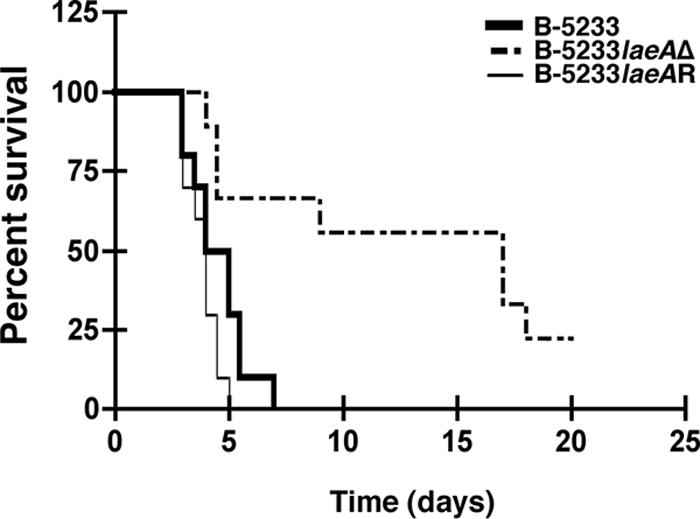
Virulence studies. Immunosuppressed 129/Sv mice were inoculated intranasally with 5 × 106 conidia. Mortality was monitored for 21 days. A P value of 0.0076 was found for comparison of survival between B-5233- and B-5233laeAΔ-infected mice. A Kaplan-Meier survival curve with a log rank test was used to compare survival levels among the strain groups.
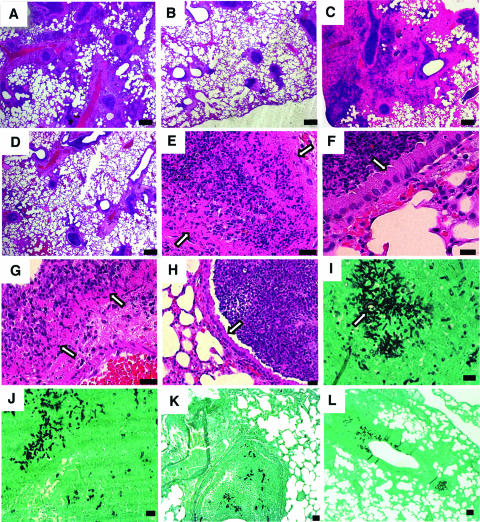
For histopathological studies, two mice infected with B-5233, B-5233laeAΔ, or B-5233laeAR were sacrificed at 72 and 96 h post-intranasal inoculation. By 72 h, lung sections from mice infected with the wild-type strain had multifocal necrotizing bronchopneumonia with neutrophilic infiltration, airways filled with cellular debris, and hyphae (Fig. 6A, E, and I). In affected bronchioles, there was full-thickness necrosis extending from the epithelium (Fig. 6E) through the surrounding smooth muscle and into the adjacent pulmonary parenchyma. Flanking the necrotic areas were small zones of hemorrhage and edema. At 72 h, rare conidial heads (Fig. 6I) along with many hyphae were seen primarily within the bronchioles of mice infected with B-5233. Similar changes were seen in mice infected with strain B-5233laeAR (data not shown). By 96 h, hyphae of the wild-type strain were scattered throughout the lung, and the pulmonary architecture was effaced by zones of necrosis and inflammation with hemorrhage and edema (Fig. 6C). The severity of the bronchopneumonia produced in the lungs of mice infected with B-5233laeAR was similar to that of the lesions produced in mice infected with the wild-type strain. The bronchiolar epithelium was necrotic in both cases by 72 h (Fig. 6G). The pulmonary lesions generated by strain B-5233laeAΔ were clearly less severe, but the character of the lesions was not appreciably different from that for the wild-type strain (Fig. 6B and D). The lesions caused by B-5233laeAΔ were more discrete, often sparing the bronchiolar epithelium, at 72 h and 96 h (Fig. 6F and H), and at 96 h, they were associated with less edema and hemorrhage (Fig. 6D). At 96 h, fungal hyphae of strain B-5233laeAΔ were still confined primarily to the bronchioles (Fig. 6K and L). In contrast, the hyphae of the wild-type strain (Fig. 6I) and B-5233laeAR (Fig. 6J) were already scattered outside of the bronchioles at 72 h. The delayed progression of B-5233laeAΔ-induced pulmonary lesions in comparison to that for the wild-type strain was corroborated by the survival data.
FIG. 6.
Histopathology of mice infected with A. fumigatus strains. (A to H) Hematoxylin-and-eosin-stained sections of lungs from mice infected intranasally with strain B-5233, B-5233laeAΔ, or B-5233laeAR. B-5233 (A) produced a more severe bronchopneumonia than B-5233laeAΔ (B) at 72 h. At 96 h, bronchopneumonia caused by strain B-5233laeAΔ (D) was still significantly less severe than that caused by strain B-5233 (C). Scale bars in panels A to D, 100 μm. At 72 h, bronchioles of mice infected with B-5233 (E) or B-5233laeAR (G) showed a completely necrotic epithelium (arrows). The epithelium of the bronchiolar walls (arrows) in the lungs of mice infected with strain B-5233laeAΔ was mostly intact at 72 h (F) as well as at 96 h (H). Scale bars in panels E to H, 10 μm. (I to L) Lung sections stained with Gomori's methenamine silver to demonstrate the extent of hyphal growth. At 72 h, hyphae of strains B-5233 (I) and B-5233laeAR (J) are seen in the lung. The arrow in panel I points to a rare conidial head of the wild-type strain within the bronchiole. Hyphae of strain B-5233laeAΔ are seen within a bronchiole at 72 h (K). At 96 h, hyphae of strain B-5233laeAΔ were still primarily within the bronchiole (L). Scale bars in panels I to L, 20 μm.
DISCUSSION
Previously it was reported that deletion of laeA in strain AF293 resulted in conidia with a smooth surface, a phenotype similar to that observed when alb1 was deleted in strain B-5233 (34). In the present study, we show that neither the laeA deletant strain of B-5233 nor strain AF293laeAΔ exhibits alterations in conidial morphology. Like those of the wild type, the conidia were bluish green and their surfaces were decorated with protrusions. Although a few exceptional conidia (<0.1%) exhibited smooth surfaces with no clear protrusions, these were observed in all strains regardless of the presence or absence of the laeA gene. Our study also showed that deletion of laeA in B-5233 as well as in AF293 caused a reduction in the expression of alb1 during mycelial growth, whereas a previous report had indicated a pronounced increase in the transcription of alb1 in the laeA deletant (3). It is possible that this difference reflects an effect of the medium used to grow the cultures. Although alb1 is expressed primarily during the conidiation stage when B-5233 is grown on a solid medium, it has been shown that the alb1 gene is also expressed in newly germinated hyphae under certain conditions (18, 43). The function of alb1 in conidial pigment synthesis is well characterized, but its function in hyphae is unknown. Whether alb1 participates in the synthesis of DHN-like melanin or in another secondary metabolite pathway during hyphal growth in AF293 is unknown. It is possible that the lack of dark color in the submerged mycelium of AF293laeAΔ is related to the down-regulation of alb1 expression in the mycelium.
The previous study with strain AF293 (3) and the present study with B-5233 showed that laeA plays a role in regulating the synthesis of gliotoxin and is also involved in the virulence of A. fumigatus in a murine model. Comparisons with B-5233 showed that B-5233laeAΔ produces 80% less gliotoxin and is slower in causing fatal infection in 129/Sv mice immunosuppressed with hydrocortisone. Furthermore, the culture filtrate of the laeA deletant is less cytotoxic to mammalian cells and has a reduced inhibitory effect on the neutrophil respiratory burst. Similarly, Bok and collaborators have reported that strain AF293laeAΔ showed significantly reduced virulence relative to AF293 in ICR mice. In addition, gliotoxin was not detected in the lungs of mice infected with AF293laeAΔ, whereas it was readily detected in the lungs of mice infected with AF293 (3). The immunosuppressive and proapoptotic properties of gliotoxin (11, 20, 29, 32) indicate that this secondary metabolite is an important factor in the pathobiology of A. fumigatus. Recent studies, however, have suggested that abrogation of gliotoxin synthesis through deletion of gliP has no effect on the virulence of A. fumigatus strain AF293 in mice immunosuppressed with cyclophosphamide and cortisone (4, 8, 15). In contrast, we observed that deletion of gliP in strain B-5233 resulted in a mutant that was significantly less virulent than the wild type in mice immunosuppressed with hydrocortisone alone (31b). This disparity is likely due to the different immunosuppressive regimens used in the murine models. Although, based on our findings, we can hypothesize that gliotoxin is an important virulence determinant of A. fumigatus, it is probable that laeA regulates the synthesis of other secondary metabolites or proteins that may also play a role in the pathobiology of this fungus. In fact, it has recently been shown that the LaeA protein regulates the expression of at least 9.5% of the A. fumigatus genome and positively regulates the expression of genes involved in the synthesis of 20 to 40% of the major classes of secondary metabolites (25). The ability of the LaeA protein to regulate the synthesis of different secondary metabolites has also been suggested to occur in Aspergillus nidulans (5).
Histological sections of lungs from mice infected with strain B-5233laeAΔ or B-5233 supported the survival data. Although the pattern of lung lesions showed no appreciable difference, the progression of the disease was clearly less severe for mice infected with the laeAΔ strain. Although B-5233laeAΔ produces conidial heads as abundantly as the wild-type strain in vitro, conidial heads with spores were found only in the bronchioles of lungs from mice infected with either B-5233 or B-5233laeAR. It is not likely that the conidial heads were part of the inoculum, since the inoculum was filtered through multiple layers of sterile gauze to eliminate hyphae and conidiophores so that it contained only free conidia. Formation of conidial heads by aspergilli in vivo has been seen in tissues where oxygen tension is high, such as in the sinus or the chronic cavitary lesions of the lung (16). It is not clear why B-5233 and B-5233laeAR, but not B-5233laeAΔ, contained conidial heads within the bronchioles. Histological examinations using more mice may be needed to find out whether this phenomenon is related to the function of the laeA gene.
The findings presented here indicate that laeA regulates the synthesis of the secondary metabolite gliotoxin and the expression of the alb1 gene during growth in liquid culture. Our study showed that deletion of laeA caused a down-regulation of alb1 expression in mycelia of both B-5233 and AF293, whereas the previous report has indicated a pronounced up-regulation of alb1 in the laeA deletant (3). It is possible that this difference reflects the media used to grow the cultures. Furthermore, such regulation of alb1 by laeΑ is apparently unrelated to the regulation of conidial morphology as revealed by SEM. This study confirms that the deletion of laeA reduces virulence in an animal model, as has been reported for AF293 (3). The reduced ability of the laeA deletant to cause apoptosis in mammalian cells and inhibition of the respiratory burst in human neutrophils may have contributed to the observed reduction in virulence.
Acknowledgments
We thank N. P. Keller, University of Wisconsin—Madison, for providing the AF293 strains and A. Varma for critical review of the manuscript.
This study was supported by funds from the intramural program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, and by a grant (Technology Transfer) from the Max Planck Society, Germany. J. Pardo was supported by the Alexander von Humboldt Foundation, Germany, and E. M. Galvez was supported by the Deutsche Forschungsgemeinschaft (DFG), Germany.
Footnotes
Published ahead of print on 13 July 2007.
REFERENCES
- 1.Amitani, R., T. Murayama, R. Nawada, W. J. Lee, A. Niimi, K. Suzuki, E. Tanaka, and F. Kuze. 1995. Aspergillus culture filtrates and sputum sols from patients with pulmonary aspergillosis cause damage to human respiratory ciliated epithelium in vitro. Eur. Respir. J. 8:1681-1687. [DOI] [PubMed] [Google Scholar]
- 2.Belkacemi, L., R. C. Barton, V. Hopwood, and E. G. Evans. 1999. Determination of optimum growth conditions for gliotoxin production by Aspergillus fumigatus and development of a novel method for gliotoxin detection. Med. Mycol. 37:227-233. [PubMed] [Google Scholar]
- 3.Bok, J. W., S. A. Balajee, K. A. Marr, D. Andes, K. F. Nielsen, J. C. Frisvad, and N. P. Keller. 2005. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 4:1574-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bok, J. W., D. Chung, S. A. Balajee, K. A. Marr, D. Andes, K. F. Nielsen, J. C. Frisvad, K. A. Kirby, and N. P. Keller. 2006. GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect. Immun. 74:6761-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bok, J. W., D. Hoffmeister, L. A. Maggio-Hall, R. Murillo, J. D. Glasner, and N. P. Keller. 2006. Genomic mining for Aspergillus natural products. Chem. Biol. 13:31-37. [DOI] [PubMed] [Google Scholar]
- 6.Bok, J. W., and N. P. Keller. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bok, J. W., D. Noordermeer, S. P. Kale, and N. P. Keller. 2006. Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol. Microbiol. 61:1636-1645. [DOI] [PubMed] [Google Scholar]
- 8.Cramer, R. A., Jr., M. P. Gamcsik, R. M. Brooking, L. K. Najvar, W. R. Kirkpatrick, T. F. Patterson, C. J. Balibar, J. R. Graybill, J. R. Perfect, S. N. Abraham, and W. J. Steinbach. 2006. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot. Cell 5:972-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denning, D. W.1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-805. [DOI] [PubMed] [Google Scholar]
- 10.Dupont, B., M. Huber, S. J. Kim, and J. E. Bennett. 1987. Galactomannan antigenemia and antigenuria in aspergillosis: studies in patients and experimentally infected rabbits. J. Infect. Dis. 155:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Eichner, R. D., M. Al Salami, P. R. Wood, and A. Mullbacher. 1986. The effect of gliotoxin upon macrophage function. Int. J. Immunopharmacol. 8:789-797. [DOI] [PubMed] [Google Scholar]
- 12.Jahn, B., A. Koch, A. Schmidt, G. Wanner, H. Gehringer, S. Bhakdi, and A. A. Brakhage. 1997. Isolation and characterization of a pigmentless-conidium mutant of Aspergillus fumigatus with altered conidial surface and reduced virulence. Infect. Immun. 65:5110-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamei, K., A. Watanabe, K. Nishimura, and M. Miyaji. 2002. Cytotoxicity of Aspergillus fumigatus culture filtrate against macrophages. Nippon Ishinkin Gakkai Zasshi 43:37-41. [DOI] [PubMed] [Google Scholar]
- 14.Keller, N., J. Bok, D. Chung, R. M. Perrin, and E. Keats Shwab. 2006. LaeA, a global regulator of Aspergillus toxins. Med. Mycol. 44(Suppl.):83-85. [DOI] [PubMed] [Google Scholar]
- 15.Kupfahl, C., T. Heinekamp, G. Geginat, T. Ruppert, A. Härtl, H. Hof, and A. A. Brakhage. 2006. Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Mol. Microbiol. 62:292-302. [DOI] [PubMed] [Google Scholar]
- 16.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology, p. 201-247. Lea & Febiger, Philadelphia, PA.
- 17.Langfelder, K., B. Jahn, H. Gehringer, A. Schmidt, G. Wanner, and A. A. Brakhage. 1998. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 187:79-89. [DOI] [PubMed] [Google Scholar]
- 18.Langfelder, K., B. Philippe, B. Jahn, J. P. Latge, and A. A. Brakhage. 2001. Differential expression of the Aspergillus fumigatus pksP gene detected in vitro and in vivo with green fluorescent protein. Infect. Immun. 69:6411-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marr, K. A., T. Patterson, and D. Denning. 2002. Aspergillosis. Pathogenesis, clinical manifestations, and therapy. Infect. Dis. Clin. N. Am. 16:875-894. [DOI] [PubMed] [Google Scholar]
- 20.Müllbacher, A., P. Waring, and R. D. Eichner. 1985. Identification of an agent in cultures of Aspergillus fumigatus displaying anti-phagocytic and immunomodulating activity in vitro. J. Gen. Microbiol. 131:1251-1258. [DOI] [PubMed] [Google Scholar]
- 21.Murayama, T., R. Amitani, Y. Ikegami, R. Nawada, W. J. Lee, and F. Kuze. 1996. Suppressive effects of Aspergillus fumigatus culture filtrates on human alveolar macrophages and polymorphonuclear leucocytes. Eur. Respir. J. 9:293-300. [DOI] [PubMed] [Google Scholar]
- 22.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 23.Pardo, J., A. Bosque, R. Brehm, R. Wallich, J. Naval, A. Mullbacher, A. Anel, and M. M. Simon. 2004. Apoptotic pathways are selectively activated by granzyme A and/or granzyme B in CTL-mediated target cell lysis. J. Cell Biol. 167:457-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardo, J., C. Urban, E. M. Galvez, P. G. Ekert, U. Muller, J. Kwon-Chung, M. Lobigs, A. Mullbacher, R. Wallich, C. Borner, and M. M. Simon. 2006. The mitochondrial protein Bak is pivotal for gliotoxin-induced apoptosis and a critical host factor of Aspergillus fumigatus virulence in mice. J. Cell Biol. 174:509-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perrin, R. M., N. D. Fedorova, J. W. Bok, R. A. Cramer, J. R. Wortman, H. S. Kim, W. C. Nierman, and N. P. Keller. 2007. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 3:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rex, J. H., J. E. Bennett, J. I. Gallin, H. L. Malech, E. S. DeCarlo, and D. A. Melnick. 1991. In vivo interferon-gamma therapy augments the in vitro ability of chronic granulomatous disease neutrophils to damage Aspergillus hyphae. J. Infect. Dis. 163:849-852. [DOI] [PubMed] [Google Scholar]
- 27.Rex, J. H., J. E. Bennett, J. I. Gallin, H. L. Malech, and D. A. Melnick. 1990. Normal and deficient neutrophils can cooperate to damage Aspergillus fumigatus hyphae. J. Infect. Dis. 162:523-528. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanzani, M., E. Orciuolo, R. Lewis, D. P. Kontoyiannis, S. L. Martins, L. S. St John, and K. V. Komanduri. 2005. Aspergillus fumigatus suppresses the human cellular immune response via gliotoxin-mediated apoptosis of monocytes. Blood 105:2258-2265. [DOI] [PubMed] [Google Scholar]
- 30.Sugareva, V., A. Hartl, M. Brock, K. Hubner, M. Rohde, T. Heinekamp, and A. A. Brakhage. 2006. Characterisation of the laccase-encoding gene abr2 of the dihydroxynaphthalene-like melanin gene cluster of Aspergillus fumigatus. Arch. Microbiol. 186:345-355. [DOI] [PubMed] [Google Scholar]
- 31.Sugui, J. A., Y. C. Chang, and K. J. Kwon-Chung. 2005. Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: an efficient tool for insertional mutagenesis and targeted gene disruption. Appl. Environ. Microbiol. 71:1798-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Sugui, J. A., J. Pardo, Y. C. Chang, K. A. Zarember, G. Nardone, E. M. Galvez, A. Müllbacher, J. I. Gallin, M. M. Simon, and K. J. Kwon-Chung. 2007. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 6:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutton, P., N. R. Newcombe, P. Waring, and A. Mullbacher. 1994. In vivo immunosuppressive activity of gliotoxin, a metabolite produced by human pathogenic fungi. Infect. Immun. 62:1192-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang, C. M., J. Cohen, T. Krausz, S. Van Noorden, and D. W. Holden. 1993. The alkaline protease of Aspergillus fumigatus is not a virulence determinant in two murine models of invasive pulmonary aspergillosis. Infect. Immun. 61:1650-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai, H. F., Y. C. Chang, R. G. Washburn, M. H. Wheeler, and K. J. Kwon-Chung. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180:3031-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai, H. F., I. Fujii, A. Watanabe, M. H. Wheeler, Y. C. Chang, Y. Yasuoka, Y. Ebizuka, and K. J. Kwon-Chung. 2001. Pentaketide melanin biosynthesis in Aspergillus fumigatus requires chain-length shortening of a heptaketide precursor. J. Biol. Chem. 276:29292-29298. [DOI] [PubMed] [Google Scholar]
- 36.Tsai, H. F., R. G. Washburn, Y. C. Chang, and K. J. Kwon-Chung. 1997. Aspergillus fumigatus arp1 modulates conidial pigmentation and complement deposition. Mol. Microbiol. 26:175-183. [DOI] [PubMed] [Google Scholar]
- 37.Tsai, H. F., M. H. Wheeler, Y. C. Chang, and K. J. Kwon-Chung. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181:6469-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsunawaki, S., L. S. Yoshida, S. Nishida, T. Kobayashi, and T. Shimoyama. 2004. Fungal metabolite gliotoxin inhibits assembly of the human respiratory burst NADPH oxidase. Infect. Immun. 72:3373-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Washburn, R. G., J. I. Gallin, and J. E. Bennett. 1987. Oxidative killing of Aspergillus fumigatus proceeds by parallel myeloperoxidase-dependent and -independent pathways. Infect. Immun. 55:2088-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei, M. C., W. X. Zong, E. H. Cheng, T. Lindsten, V. Panoutsakopoulou, A. J. Ross, K. A. Roth, G. R. MacGregor, C. B. Thompson, and S. J. Korsmeyer. 2001. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292:727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida, L. S., S. Abe, and S. Tsunawaki. 2000. Fungal gliotoxin targets the onset of superoxide-generating NADPH oxidase of human neutrophils. Biochem. Biophys. Res. Commun. 268:716-723. [DOI] [PubMed] [Google Scholar]
- 42.Zarember, K. A., J. A. Sugui, Y. C. Chang, K. J. Kwon-Chung, and J. I. Gallin. 2007. Human PMN inhibit Aspergillus fumigatus conidial growth by lactoferrin-mediated iron depletion. J. Immunol. 178:6367-6373. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, L., M. Wang, R. Li, and R. Calderone. 2005. Expression of Aspergillus fumigatus virulence-related genes detected in vitro and in vivo with competitive RT-PCR. Mycopathologia 160:201-206. [DOI] [PubMed] [Google Scholar]