Introduction
Globoid cell leukodystrophy (GLD), or Krabbe disease, is an autosomal recessive disorder caused by decreased ceramide beta-galactosidase (GALC) activity (Schmitteckert, et al., 1999). To date, more than 60 mutations causing the disease in humans have been reported (Wenger, et al., 2000;Escolar, et al., 2005). The disease is characterized by oligodendrocyte degeneration and apoptosis causing demyelination of the central and peripheral nervous system. In infants, GLD manifests as irritability, spasticity, blindness, deafness, seizures, and impaired cognitive and motor development. GLD progresses rapidly and generally results in death before two years of age, although there is also a less common, late onset form in adults (Wenger, et al., 2000). Bone marrow or umbilical-cord blood transplantation has been shown to prevent disease, but is only effective if given prior to the onset of symptoms (Escolar, et al., 2005). GALC deficiency has been documented in several other species, and three main animal models of GLD are utilized: canine, murine, and simian. The twitcher mouse is the most widely used of these models, owing to a high degree of genetic manipulability and the comparatively low cost of breeding and maintenance. The rhesus macaque model is the only nonhuman primate model of a genetic disorder, and macaque GALC shares 97% homology with the human gene (Luzi, et al., 1997). The rhesus macaque model is housed at the Tulane National Primate Research Center in Covington, Louisiana, where a colony of 60 heterozygous carriers is bred and maintained (Baskin, et al., 1998).
Animal models of globoid cell leukodystrophy have significantly advanced our understanding of the pathogenesis of the disease and allow testing of therapeutic strategies (Wenger, 2000). In recent years, the twitcher mouse model has been used to test the effectiveness and mechanism of peripheral enzyme replacement therapy (Umezawa, et al., 1985;Lee, et al., 2005), bone marrow transplantation (Yeager, et al., 1984;Seller, et al., 1986;Ichioka, et al., 1987;Hoogerbrugge, et al., 1988a;Hoogerbrugge, et al., 1988b;Kondo, et al., 1988;Suzuki, et al., 1988;Yeager, et al., 1991;Wu, et al., 2001;Biswas, LeVine, 2002;Yagi, et al., 2004), cell transplantation therapy (Scaravilli, Jacobs, 1981;Huppes, et al., 1992;Croitoru-Lamoury, et al., 2006;Pellegatta, et al., 2006;Taylor, et al., 2006), and viral vector gene therapy (Luddi, et al., 2001;Shen, et al., 2001;Lin, et al., 2005;Shen, et al., 2005;Dolcetta, et al., 2006;Lin, et al., 2007). It has also been used to study molecular pathogenetic mechanisms of lipid metabolism (Kobayashi, et al., 1986;Kobayashi, et al., 1987;Mitsuo, et al., 1989;Taniike, et al., 1998;Ezoe, et al., 2000;LeVine, et al., 2000;Biswas, et al., 2003;Esch, et al., 2003), effects of inflammatory mediators on progression of disease (Matsushima, et al., 1994;LeVine, Brown, 1997;Taniike, et al., 1997;Pedchenko, LeVine, 1999;Pedchenko, et al., 2000;Biswas, et al., 2001;Mohri, et al., 2006), alterations in cellular signaling cascades (Yamada, et al., 1996;Yamada, Suzuki, 1999;Giri, et al., 2006), and alterations in peroxisomal function (Khan, et al., 2005;Haq, et al., 2006). The rhesus macaque model has been described (Luzi, et al., 1997;Baskin, et al., 1998;Weimer, et al., 2005) and its use will be critical for the safe translation of therapies in animal models into the human population.
The simian and murine models of GLD are due to well-defined mutations in the GALC gene, resulting in premature termination of transcription and formation of an enzymatically inactive protein fragment. The twitcher mutation is a single nucleotide substitution which results in the formation of a stop codon (Kobayashi, et al., 1980;Sakai, et al., 1996). In the rhesus macaque, a 2 basepair deletion causes a frame shift during transcription and a premature stop codon 48 nucleotides downstream from the mutation (Luzi, et al., 1997). Restriction enzyme (RE) digest analysis has traditionally been used in genotyping these animals. However, RE digest is a time-consuming and labor-intensive process which sometimes produces an ambiguous result. Here we present a rapid, high-throughput, unambiguous molecular beacon assay for both mouse and rhesus models of GLD which is an improvement over RE digestion genotyping. We demonstrate that this assay can be used to distinguish between affected, wild-type, and heterozygous individuals from hair root samples in under 4 hours. This procedure can be performed using a standard thermocycler with a 96-well plate block and a fluorescent plate reader; a real-time thermocycler is not required.
Materials and Methods
Sample Preparation
Allelic DNA standards were generated from tissues of known genotype by proteinase K digestion followed by phenol/chloroform extraction and ethanol precipitation. DNA was extracted from hair root samples of undetermined genotype following the Chelex-100® protocol described by Walsh (Walsh, et al., 1991). Briefly, hair roots were added to 100 μl of 10% (w/v) Chelex-100® 200-400 mesh (Bio-Rad, Hercules, CA, USA) solution. DNA was polymerase chain reaction (PCR) amplified from entire hair strands and even a single hair root; however optimal amplification was achieved using 20 - 40 hair roots. The samples were incubated at 100°C for 20 minutes with periodic vortexing. After brief centrifugation, the supernatant was removed and combined with 2 μl of 1 M Tris-HCl pH 8.0 to reduce alkalysis of DNA during storage. Samples were immediately used for PCR or were stored at -20°C.
Rhesus macaque DNA was also amplified directly from whole blood stored on Whatman FTA® Cards (VWR Scientific, Suwanee, GA, USA). 1.2 mm discs were punched from the cards and prepared for PCR following the manufacturer’s protocol. Discs were removed from PCR tubes prior to fluorescence analysis.
Polymerase Chain Reaction
Each 20 μl amplification reaction contained 20 mM Tris-HCl pH 8.4, 50 mM KCl, 4 mM MgCl2, 0.2 mM each dNTP, 0.4μM of each corresponding forward and reverse primer, 0.25 μM mutant beacon, 0.25 μM wild-type beacon, 0.04 U/μl of Platinum Taq (Invitrogen, Carlsbad, CA, USA), and template DNA. Template DNA was in the form of 2 μl Chelex-100® extract, one 1.2 mm Whatman FTA® Card disc, or 5-20 ng of allelic standard DNA. Samples were amplified in an MJ Research PTC-100 thermocycler by 40 cycles of denaturation at 95°C for 15 s, annealing at 50°C/45°C (mouse/macaque, respectively) for 15 s, and extension at 72°C for 15 s. In some cases, fewer cycles were run to determine ratio linearity. No-template controls were included in each run. Molecular beacons were designed using Beacon Designer 4 (Premier Biosoft International, Palo Alto, CA, USA) and purchased from Integrated DNA Technologies (Coralville, IA, USA). Primers and beacons (Table I) were designed based on the GALC gene sequences as listed in Genbank (mouse accession no. NC_000078, macaque accession no. U87464). For both assays, wild-type beacons were conjugated at the 5’ end to hexachlorofluorescein (HEX™) which demonstrated maximum emission at 560 nm, while mutant beacons were conjugated at the 5’ end to fluorescein (6-FAM) which demonstrated maximum emission at 535 nm. All beacons were conjugated at the 3’ end to the quencher molecule Dabcyl. Predicted conformation of the molecular beacons is shown in hairpin and bound forms in Figure 1.
Table I.
| Twitcher Mouse | ||
| Molecular Beacon PCR | ||
| Forward primer | 5′-CTG CTT AGA ATC AAT CAG ACT G | -3′ |
| Reverse primer | 5′-CTC AAC AAC GGA CAA TTA CC | -3′ |
| Wild type beacon | 5′-[HEX]CCA GGC TGG TAT TAC CTG G[Dabcyl] | -3′ |
| Mutant beacon | 5′-[FAM]CCA GGC TGA TAT TAC CTG G[Dabcyl] | -3′ |
| Restriction Digest PCR | ||
| gTwF | 5′-CAC TTA ATT TTC TCC AGT CAT | -3′ |
| TwR | 5′-TAG ATG GCC CAC TGT CTT CAG GTG ATA | -3′ |
| Rhesus Macaque | ||
| Molecular Beacon PCR | ||
| Forward primer | 5′-TGG TTA ATG AAG GAA GC | -3′ |
| Reverse primer | 5′-GAT TCC ACC AAC ACG A | -3′ |
| Wild type beacon | 5′-[HEX]CCC AAT ATT ACA CTC ATT GGG[Dabcyl] | -3′ |
| Mutant beacon | 5′-[FAM]CCC AAT ATT ACT CAT TGG G[Dabcyl] | -3′ |
| Restriction Digest PCR | ||
| rhGALCF | 5′-AGA AGA GGA ATC CCA ATA GTA C | -3′ |
| rhGALCR | 5′-CCA TAC TAA TAG AGA TTC CAC CA | -3′ |
Figure 1.

Molecular beacon design for mouse (A) and rhesus macaque (B) GALC genes. The mutant beacon (top left) binds to the antisense strand of the mutant genomic DNA allowing the detection of fluorescence (bottom center), while the wild-type beacon remains in a closed hairpin formation in which fluorescence is quenched (top right). Boxed nucleotides show sites of single nucleotide substitution in mouse and two base deletion (arrowheads) in rhesus macaque. Note the natural palindromic structure of the GALC genes, which was incorporated into the beacon sequences.
Real-Time Fluorescence Analysis
Fluorescence was measured after completion of PCR using an ABI7700 Real-Time Sequence Detection System (Applied Biosystems, Inc., Foster City, CA, USA) over a spectrum of 500 nm - 600 nm with an excitation wavelength of 480 nm. Values were recorded at 25°C for genotype determination and at 95°C to ensure the presence and functioning of both HEX™ and FAM fluorophores. Optimal exposure time was found to be 25 ms, however accurate readings were also obtained from 15 ms exposures. Samples were easily genotyped by direct comparison of spectral shape (Figure 2) or by plotting 535nm values against 560 nm values on standard axes (Figure 3). Beacon melting temperature analysis was performed by using the ABI7700 to monitor fluorescence as temperature decreased from 95°C to 25°C.
Figure 2.

Genotype determination of an infant rhesus macaque using the molecular beacon assay. The fluorescence emission spectrum is shown from 500 nm to 600 nm for each of the genotype standards as well as the infant′s DNA, extracted from hair roots. Comparison of the unknown sample to each standard clearly indicates that the infant is heterozygous for the GALC mutation. Fluorescence is expressed in arbitrary units. Red is Krabbe standard, green is heterozygous standard, blue is wild-type standard, purple is infant hair root DNA, and black is no template control.
Figure 3.
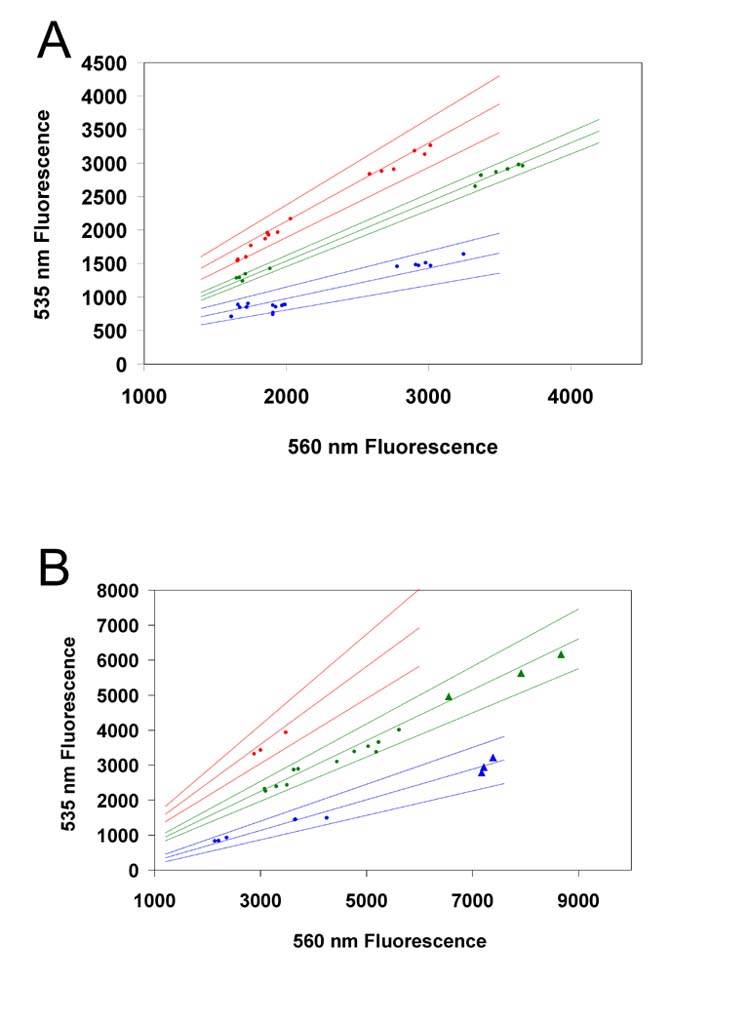
High-throughput genotyping of mice (A) and rhesus macaques (B) based on 535nm/560nm fluorescence ratios. For clarity, homozygous mutants are shown in red, heterozygous in yellow, and homozygous wild-type in blue. Hair samples are represented as dots and blood card samples as triangles. Regressions are shown for each genotype with upper and lower 99% confidence limits. Blood card samples were only tested for rhesus macaques; a blood card sample was not available for the homozygous mutant monkey. The number of standards used to generate the confidence limits were (A) 26 wild type, 24 heterozygous, 23 homozygous mutant, (B) 9 wild type, 9 heterozygous, 9 homozygous mutant.
Plate-Reader Fluorescence Analysis
PCR was performed using a black well plate (Island Scientific, Bainbridge Island, WA, USA) in an MJ Research PTC-100 thermocycler. Fluorescence was read using a Fluoroskan Ascent plate reader (Thermo Electron Corp/Labsystems, Waltham, MA, USA) with 435 nm excitation and 538 nm (mutant) or 555 nm (wild type) emission and a 200 msec integration time per well.
Restriction Enzyme Digest
Restriction enzyme digests of murine PCR-amplified samples were performed following published methods (Sakai, et al., 1996). PCR conditions were identical to those used in the molecular beacon assays except that the MgCl2 concentration was 2 mM and molecular beacons were omitted. Primers gTwF, which is mismatched from template DNA for the purpose of generating part of the EcoRV site, and TwR were used with annealing at 47°C. Amplified standards and hair root samples of each genotype were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA, USA) and digested with EcoRV (Invitrogen) in the supplied buffer for 4 hours at 37°C. The digest was separated by 12% polyacrylamide gel electrophoresis at 40 volts for 6 hours. Gels were stained in 0.5 μg/ml ethidium bromide (Sigma, St. Louis, MO, USA) and imaged using a Quantity-One gel documentation system (Bio-Rad).
Rhesus macaque RE digests were performed according to published methods (Luzi, et al., 1997). PCR conditions were identical to those used for the murine RE digests, with the exception that rhGALCF and rhGALCR primers were used with an annealing temperature of 45°C. Purified PCR product was digested with ScaI (Invitrogen) in the supplied buffer for 4 hours at 37°C.
Statistical Analysis
Fluorescence 535/560 nm ratios were tested for significant differences between genotype using standard one-way ANOVA, followed by Tukey′s Studentized Range test for inter-genotype comparisons. Linear regression of fluorescence emission data used Microsoft Excel (Microsoft, Redmond, WA, USA) and 99% confidence intervals were plotted using the regression y-intercept. Finally, Student′s t test was performed between standard and sample ratios within each genotype to establish that samples were not significantly different than standards. Residual by rank and residual by predicted ratio plots were used to establish normality and constant variance of ratio data. Software analysis of data was performed with SAS software version 9.1 for the PC (SAS Institute Inc., Cary, NC, USA).
Results
The melting temperatures of the molecular beacon in PCR buffer without DNA template were 64°C and 59°C for the mouse wild type and twitcher mutant beacons, and 58°C and 60°C for the rhesus macaque wild type and mutant beacons (data not shown). The predicted hairpin and bound structures are given in Figure 1. Restriction enzyme digestion of DNA standards and Chelex-100® hair root extracts yielded expected band sizes. For the mouse, the amplified fragment was 260 bp in length, and mutant amplification products were cut with EcoRV into 234 and 26 bp fragments (Figure 4A). For the rhesus macaque, the amplified fragment was 120 bp in length, and mutant amplification products were cut with ScaI into 20 and 98 bp fragments (Figure 4B).
Figure 4.
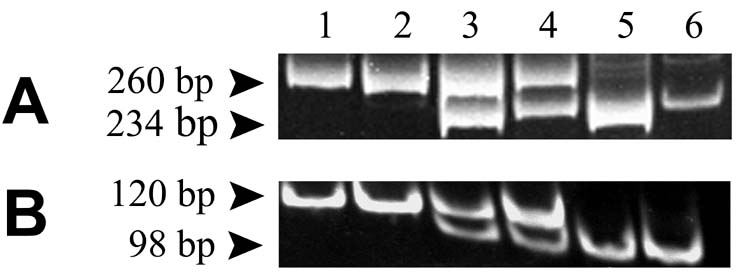
Restriction enzyme digestion for genotype determination of mouse (A) and rhesus macaque (B). PCR products were digested with EcoRV and ScaI and separated on 12% polyacrylamide gels with detection by ethidium fluorescence. In each case, the mutant allele generates a cut site when amplified with appropriate primers listed in Table I. Lanes are 1, wild-type standard; 2, wild-type sample; 3, heterozygous standard; 4, heterozygous sample; 5, homozygous mutant standard; 6, homozygous mutant sample.
The emission spectra of HEX™ and FAM probes overlap; however the spectral emission curves generated by the molecular beacon assay are distinct by genotype. The average 535 and 560 nm fluorescence values from three reactions without template were subtracted from each of the samples prior to 535/560 nm ratio determination. For both the mouse and the rhesus macaque, homozygous mutant samples result in 535 nm emission exceeding 560 nm emission, heterozygous samples result in 535 nm emission approximately half as intense as in the mutant, and homozygous wild type samples give 535 nm emission near baseline. The 560 nm emission remains relatively constant in multiplex reactions due to approximately equal fluorescence of FAM and HEX™ fluorophores at this wavelength. In the absence of the mutant (FAM) beacon, the wild type (HEX™) beacon fluoresces at 560 nm only in the presence of the wild type allele (data not shown).
Pooled analysis of multiple runs using the molecular beacon assay shows constant 535/560 nm emission ratios for each genotype by species regardless of the level of amplification or source of specimen, which is presented in Figure 3 as the linear regression of the 535/560 nm slope. The 99% confidence interval of the linear regression of standard DNA samples shows no overlap for the range of beacon fluorescence generated by modifying the standard concentration, the number of cycles of amplification and ABI Prism exposure times.
Table II presents 538/555 nm fluorescence emission intensity ratio data for the molecular beacon assays determined by a fluorescent plate reader (n=3 for all ratios). The standard ratio ranges of the mean ± 3 standard deviations for each genotype standard ratio do not overlap and contain the hair root sample ratios. Fluorescence emission intensity ratios were significantly different between genotypes as determined by ANOVA (F=9101, p < 0.001) followed by Tukey′s studentized range test for pairwise comparison between groups (t=2.13, p < 0.05). Mean fluorescence emission ratios were not significantly different between standards and samples of identical genotype as determined by Student′s t test (p > 0.1).
Table II.
| Standard 538nm/555nm (mean ± std dev) | Sample 538nm/555nm (mean ± std dev) | |
|---|---|---|
| Twitcher Mouse | ||
| Homozygous mutant | 2.068 ± 0.025 | 2.066 ± 0.025 |
| Heterozygous | 1.525 ± 0.023 | 1.496 ± 0.033 |
| Homozygous wild-type | 0.630 ± 0.014 | 0.608 ± 0.036 |
| Rhesus Macaque | ||
| Homozygous mutant | 2.099 ± 0.016 | 2.119 ± 0.007 |
| Heterozygous | 1.306 ± 0.026 | 1.285 ± 0.029 |
| Homozygous wild-type | 0.666 ± 0.001 | 0.674 ± 0.009 |
We tested the effect of intentional sample contamination with hairs of differing genotype on the 538/555 nm fluorescence ratio. We found that including 4-10 hairs of differing genotype in an extraction sample of 25 hairs caused the ratio to vary from each standard by greater than 20% of the standard, whereas noncontaminated samples vary from purified DNA standard by less than 10% (data not shown). In practice, we find it quite easy to avoid sample contamination since hairs are readily visible and easily cleaned from instruments with a damp wiper.
Discussion
The molecular beacon assays presented here constitute a novel approach to genotyping murine and rhesus macaque models of globoid cell leukodystrophy. The GALC mutation has conventionally been detected in these models by restriction enzyme digest of the amplified gene segment or by enzymatic activity assay; however, these methods are time consuming and sometimes yield ambiguous results. This pair of molecular beacon assays reduce the procedure to a single step DNA extraction and PCR with immediate fluorescence analysis. These molecular beacon assays reliably discriminate among nearly identical alleles. Because the beacon must have a strong affinity for the template to abandon its own stable hairpin structure, a single base pair mismatch is sufficient to markedly reduce binding and fluorescence. In the mutant macaque GALC gene the beacons distinguish a two-nucleotide deletion, whereas in the mouse a single G to A point mutation is distinguished. In both of these models the sequence of the GALC gene presented a challenge to the design of an effective beacon. The murine and macaque GALC genes have palindromic sequences flanking the mutation sites which cause hairpin formation of the probe in competition with binding to amplified target sequences. We incorporated these palindromic regions into the stem portion of the beacon to generate a beacon that binds amplified target DNA along nearly its entire length (Figure 1).
Several years ago, the use of hair root DNA as an alternative to screen transgenic animals was presented by Schmitteckert and colleagues (Schmitteckert, et al., 1999). However, despite the animal welfare advantages and time and cost savings, this method has still not attained widespread use. Hair roots permit sampling of many mice very quickly because tail snipping often requires cauterization for hemostasis. Additionally, sampling hair roots from rhesus macaques avoids unnecessary exposure to sharps and bloodborne pathogens, and permits sampling in very young monkeys in which venipuncture is difficult. We have also successfully genotyped neonatal macaques from DNA extracted from chorionic villus biopsies 1 mm in size (data not shown), and adults from blood-soaked filter paper (FTA Blood cards, Whatman). The use of blood-soaked filter paper to generate a genomic data bank for an entire outbred breeding colony could allow rapid genotyping of thousands of individuals as new assays are developed. The molecular beacon assay reagents cost approximately $0.94/reaction compared to $1.51/reaction for the conventional method, not including technician time or electrophoresis cost. Furthermore, we have found that the molecular beacon assay has been in concordance with replicates and with restriction enzyme digestion results on every occasion.
Molecular beacon assays provide an ideal method by which to quickly and accurately genotype models of genetic disease. The murine and rhesus macaque assays that we have developed for the detection of the GALC mutation have consistently provided unambiguous results and clearly surpass conventional methods of genotyping these models. When combined with rapid DNA extraction methods, such as Chelex-100® or blood-soaked filter paper detergent extraction, reliable genotyping can be carried out in only a few hours. This facilitates high-throughput genotyping and is valuable whenever efficient, reliable genotyping is essential.
Acknowledgements
We gratefully acknowledge funding support from NIH grants RR00154, NS30769, RR00164, and RR022826, and assistance of the animal care support staff of the Tulane National Primate Research Center and the Tulane University Health Sciences Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baskin GB, Ratterree M, Davison BB, Falkenstein KP, Clarke MR, England JD, Vanier MT, Luzi P, Rafi MA, Wenger DA. Genetic galactocerebrosidase deficiency (globoid cell leukodystrophy, Krabbe disease) in rhesus monkeys (Macaca mulatta) Lab Anim Sci. 1998;48:476–82. [PubMed] [Google Scholar]
- Biswas S, Biesiada H, Williams TD, LeVine SM. Substrate reduction intervention by L-cycloserine in twitcher mice (globoid cell leukodystrophy) on a B6;CAST/Ei background. Neurosci Lett. 2003;347:33–6. doi: 10.1016/s0304-3940(03)00633-5. [DOI] [PubMed] [Google Scholar]
- Biswas S, LeVine SM. Substrate-reduction therapy enhances the benefits of bone marrow transplantation in young mice with globoid cell leukodystrophy. Pediatr Res. 2002;51:40–7. doi: 10.1203/00006450-200201000-00009. [DOI] [PubMed] [Google Scholar]
- Biswas S, Pinson DM, Bronshteyn IG, LeVine SM. IL-6 deficiency allows for enhanced therapeutic value after bone marrow transplantation across a minor histocompatibility barrier in the twitcher (globoid cell leukodystrophy) mouse. J Neurosci Res. 2001;65:298–307. doi: 10.1002/jnr.1154. [DOI] [PubMed] [Google Scholar]
- Croitoru-Lamoury J, Williams KR, Lamoury FM, Veas LA, Ajami B, Taylor RM, Brew BJ. Neural transplantation of human MSC and NT2 cells in the twitcher mouse model. Cytotherapy. 2006;8:445–58. doi: 10.1080/14653240600879152. [DOI] [PubMed] [Google Scholar]
- Dolcetta D, Perani L, Givogri MI, Galbiati F, Amadio S, Del Carro U, Finocchiaro G, Fanzani A, Marchesini S, Naldini L, Roncarolo MG, Bongarzone E. Design and optimization of lentiviral vectors for transfer of GALC expression in Twitcher brain. J Gene Med. 2006;8:962–71. doi: 10.1002/jgm.924. [DOI] [PubMed] [Google Scholar]
- Esch SW, Williams TD, Biswas S, Chakrabarty A, Levine SM. Sphingolipid profile in the CNS of the twitcher (globoid cell leukodystrophy) mouse: a lipidomics approach. Cell Mol Biol (Noisy-le-grand) 2003;49:779–87. [PubMed] [Google Scholar]
- Escolar ML, Poe MD, Provenzale JM, Richards KC, Allison J, Wood S, Wenger DA, Pietryga D, Wall D, Champagne M, Morse R, Krivit W, Kurtzberg J. Transplantation of umbilical-cord blood in babies with infantile Krabbe′s disease. N Engl J Med. 2005;352:2069–81. doi: 10.1056/NEJMoa042604. [DOI] [PubMed] [Google Scholar]
- Ezoe T, Vanier MT, Oya Y, Popko B, Tohyama J, Matsuda J, Suzuki K, Suzuki K. Biochemistry and neuropathology of mice doubly deficient in synthesis and degradation of galactosylceramide. J Neurosci Res. 2000;59:170–8. doi: 10.1002/(sici)1097-4547(20000115)59:2<170::aid-jnr3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Giri S, Khan M, Rattan R, Singh I, Singh AK. Krabbe disease: psychosine-mediated activation of phospholipase A2 in oligodendrocyte cell death. J Lipid Res. 2006;47:1478–92. doi: 10.1194/jlr.M600084-JLR200. [DOI] [PubMed] [Google Scholar]
- Haq E, Contreras MA, Giri S, Singh I, Singh AK. Dysfunction of peroxisomes in twitcher mice brain: a possible mechanism of psychosine-induced disease. Biochem Biophys Res Commun. 2006;343:229–38. doi: 10.1016/j.bbrc.2006.02.131. [DOI] [PubMed] [Google Scholar]
- Hoogerbrugge PM, Poorthuis BJ, Romme AE, van de Kamp JJ, Wagemaker G, van Bekkum DW. Effect of bone marrow transplantation on enzyme levels and clinical course in the neurologically affected twitcher mouse. J Clin Invest. 1988a;81:1790–4. doi: 10.1172/JCI113521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogerbrugge PM, Suzuki K, Suzuki K, Poorthuis BJ, Kobayashi T, Wagemaker G, van Bekkum DW. Donor-derived cells in the central nervous system of twitcher mice after bone marrow transplantation. Science. 1988b;239:1035–8. doi: 10.1126/science.3278379. [DOI] [PubMed] [Google Scholar]
- Huppes W, De Groot CJ, Ostendorf RH, Bauman JG, Gossen JA, Smit V, Vijg J, Dijkstra CD. Detection of migrated allogeneic oligodendrocytes throughout the central nervous system of the galactocerebrosidase-deficient twitcher mouse. J Neurocytol. 1992;21:129–36. doi: 10.1007/BF01189011. [DOI] [PubMed] [Google Scholar]
- Ichioka T, Kishimoto Y, Brennan S, Santos GW, Yeager AM. Hematopoietic cell transplantation in murine globoid cell leukodystrophy (the twitcher mouse): effects on levels of galactosylceramidase, psychosine, and galactocerebrosides. Proc Natl Acad Sci U S A. 1987;84:4259–63. doi: 10.1073/pnas.84.12.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Haq E, Giri S, Singh I, Singh AK. Peroxisomal participation in psychosine-mediated toxicity: implications for Krabbe′s disease. J Neurosci Res. 2005;80:845–54. doi: 10.1002/jnr.20529. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Shinnoh N, Kuroiwa Y. Metabolism of galactosylceramide in the twitcher mouse, an animal model of human globoid cell leukodystrophy. Biochim Biophys Acta. 1986;879:215–20. [PubMed] [Google Scholar]
- Kobayashi T, Shinoda H, Goto I, Yamanaka T, Suzuki Y. Globoid cell leukodystrophy is a generalized galactosylsphingosine (psychosine) storage disease. Biochem Biophys Res Commun. 1987;144:41–6. doi: 10.1016/s0006-291x(87)80472-2. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Yamanaka T, Jacobs JM, Teixeira F, Suzuki K. The Twitcher mouse: an enzymatically authentic model of human globoid cell leukodystrophy (Krabbe disease) Brain Res. 1980;202:479–83. doi: 10.1016/0006-8993(80)90159-6. [DOI] [PubMed] [Google Scholar]
- Kondo A, Hoogerbrugge PM, Suzuki K, Poorthuis BJ, Van Bekkum DW, Suzuki K. Pathology of the peripheral nerve in the twitcher mouse following bone marrow transplantation. Brain Res. 1988;460:178–83. doi: 10.1016/0006-8993(88)91220-6. [DOI] [PubMed] [Google Scholar]
- Lee WC, Courtenay A, Troendle FJ, Stallings-Mann ML, Dickey CA, DeLucia MW, Dickson DW, Eckman CB. Enzyme replacement therapy results in substantial improvements in early clinical phenotype in a mouse model of globoid cell leukodystrophy. Faseb J. 2005;19:1549–51. doi: 10.1096/fj.05-3826fje. [DOI] [PubMed] [Google Scholar]
- LeVine SM, Brown DC. IL-6 and TNFalpha expression in brains of twitcher, quaking and normal mice. J Neuroimmunol. 1997;73:47–56. doi: 10.1016/s0165-5728(96)00166-x. [DOI] [PubMed] [Google Scholar]
- LeVine SM, Pedchenko TV, Bronshteyn IG, Pinson DM. L-cycloserine slows the clinical and pathological course in mice with globoid cell leukodystrophy (twitcher mice) J Neurosci Res. 2000;60:231–6. doi: 10.1002/(SICI)1097-4547(20000415)60:2<231::AID-JNR12>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Lin D, Donsante A, Macauley S, Levy B, Vogler C, Sands MS. Central Nervous System-directed AAV2/5-Mediated Gene Therapy Synergizes with Bone Marrow Transplantation in the Murine Model of Globoid-cell Leukodystrophy. Mol Ther. 2007;15:44–52. doi: 10.1038/sj.mt.6300026. [DOI] [PubMed] [Google Scholar]
- Lin D, Fantz CR, Levy B, Rafi MA, Vogler C, Wenger DA, Sands MS. AAV2/5 vector expressing galactocerebrosidase ameliorates CNS disease in the murine model of globoid-cell leukodystrophy more efficiently than AAV2. Mol Ther. 2005;12:422–30. doi: 10.1016/j.ymthe.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Luddi A, Volterrani M, Strazza M, Smorlesi A, Rafi MA, Datto J, Wenger DA, Costantino-Ceccarini E. Retrovirus-mediated gene transfer and galactocerebrosidase uptake into twitcher glial cells results in appropriate localization and phenotype correction. Neurobiol Dis. 2001;8:600–10. doi: 10.1006/nbdi.2001.0407. [DOI] [PubMed] [Google Scholar]
- Luzi P, Rafi MA, Victoria T, Baskin GB, Wenger DA. Characterization of the rhesus monkey galactocerebrosidase (GALC) cDNA and gene and identification of the mutation causing globoid cell leukodystrophy (Krabbe disease) in this primate. Genomics. 1997;42:319–24. doi: 10.1006/geno.1997.4744. [DOI] [PubMed] [Google Scholar]
- Matsushima GK, Taniike M, Glimcher LH, Grusby MJ, Frelinger JA, Suzuki K, Ting JP. Absence of MHC class II molecules reduces CNS demyelination, microglial/macrophage infiltration, and twitching in murine globoid cell leukodystrophy. Cell. 1994;78:645–56. doi: 10.1016/0092-8674(94)90529-0. [DOI] [PubMed] [Google Scholar]
- Mitsuo K, Kobayashi T, Shinnoh N, Goto I. Biosynthesis of galactosylsphingosine (psychosine) in the twitcher mouse. Neurochem Res. 1989;14:899–903. doi: 10.1007/BF00964821. [DOI] [PubMed] [Google Scholar]
- Mohri I, Taniike M, Taniguchi H, Kanekiyo T, Aritake K, Inui T, Fukumoto N, Eguchi N, Kushi A, Sasai H, Kanaoka Y, Ozono K, Narumiya S, Suzuki K, Urade Y. Prostaglandin D2-mediated microglia/astrocyte interaction enhances astrogliosis and demyelination in twitcher. J Neurosci. 2006;26:4383–93. doi: 10.1523/JNEUROSCI.4531-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedchenko TV, Bronshteyn IG, LeVine SM. TNF-receptor 1 deficiency fails to alter the clinical and pathological course in mice with globoid cell leukodystrophy (twitcher mice) but affords protection following LPS challenge. J Neuroimmunol. 2000;110:186–94. doi: 10.1016/s0165-5728(00)00345-3. [DOI] [PubMed] [Google Scholar]
- Pedchenko TV, LeVine SM. IL-6 deficiency causes enhanced pathology in Twitcher (globoid cell leukodystrophy) mice. Exp Neurol. 1999;158:459–68. doi: 10.1006/exnr.1999.7125. [DOI] [PubMed] [Google Scholar]
- Pellegatta S, Tunici P, Poliani PL, Dolcetta D, Cajola L, Colombelli C, Ciusani E, Di Donato S, Finocchiaro G. The therapeutic potential of neural stem/progenitor cells in murine globoid cell leukodystrophy is conditioned by macrophage/microglia activation. Neurobiol Dis. 2006;21:314–23. doi: 10.1016/j.nbd.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Sakai N, Inui K, Tatsumi N, Fukushima H, Nishigaki T, Taniike M, Nishimoto J, Tsukamoto H, Yanagihara I, Ozono K, Okada S. Molecular cloning and expression of cDNA for murine galactocerebrosidase and mutation analysis of the twitcher mouse, a model of Krabbe′s disease. J Neurochem. 1996;66:1118–24. doi: 10.1046/j.1471-4159.1996.66031118.x. [DOI] [PubMed] [Google Scholar]
- Scaravilli F, Jacobs JM. Peripheral nerve grafts in hereditary leukodystrophic mutant mice (twitcher) Nature. 1981;290:56–8. doi: 10.1038/290056a0. [DOI] [PubMed] [Google Scholar]
- Schmitteckert EM, Prokop CM, Hedrich HJ. DNA detection in hair of transgenic mice--a simple technique minimizing the distress on the animals. Lab Anim. 1999;33:385–9. doi: 10.1258/002367799780487922. [DOI] [PubMed] [Google Scholar]
- Seller MJ, Perkins KJ, Fensom AH. Galactosylcerebrosidase activity in tissues of twitcher mice with and without bone marrow transplantation. J Inherit Metab Dis. 1986;9:234–8. doi: 10.1007/BF01799653. [DOI] [PubMed] [Google Scholar]
- Shen JS, Meng XL, Yokoo T, Sakurai K, Watabe K, Ohashi T, Eto Y. Widespread and highly persistent gene transfer to the CNS by retrovirus vector in utero: implication for gene therapy to Krabbe disease. J Gene Med. 2005;7:540–51. doi: 10.1002/jgm.719. [DOI] [PubMed] [Google Scholar]
- Shen JS, Watabe K, Ohashi T, Eto Y. Intraventricular administration of recombinant adenovirus to neonatal twitcher mouse leads to clinicopathological improvements. Gene Ther. 2001;8:1081–7. doi: 10.1038/sj.gt.3301495. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Hoogerbrugge PM, Poorthuis BJ, Bekkum DW, Suzuki K. The twitcher mouse. Central nervous system pathology after bone marrow transplantation. Lab Invest. 1988;58:302–9. [PubMed] [Google Scholar]
- Taniike M, Marcus JR, Nishigaki T, Fujita N, Popko B, Suzuki K. Suppressed UDP-galactose: ceramide galactosyltransferase and myelin protein mRNA in twitcher mouse brain. J Neurosci Res. 1998;51:536–40. doi: 10.1002/(SICI)1097-4547(19980215)51:4<536::AID-JNR13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Taniike M, Marcus JR, Popko B, Suzuki K. Expression of major histocompatibility complex class I antigens in the demyelinating twitcher CNS and PNS. J Neurosci Res. 1997;47:539–46. [PubMed] [Google Scholar]
- Taylor RM, Lee JP, Palacino JJ, Bower KA, Li J, Vanier MT, Wenger DA, Sidman RL, Snyder EY. Intrinsic resistance of neural stem cells to toxic metabolites may make them well suited for cell non-autonomous disorders: evidence from a mouse model of Krabbe leukodystrophy. J Neurochem. 2006;97:1585–99. doi: 10.1111/j.1471-4159.2006.03986.x. [DOI] [PubMed] [Google Scholar]
- Umezawa F, Eto Y, Tokoro T, Ito F, Maekawa K. Enzyme replacement with liposomes containing beta-galactosidase from Charonia lumpas in murine globoid cell leukodystrophy (twitcher) Biochem Biophys Res Commun. 1985;127:663–7. doi: 10.1016/s0006-291x(85)80212-6. [DOI] [PubMed] [Google Scholar]
- Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–13. [PubMed] [Google Scholar]
- Weimer MB, Gutierrez A, Baskin GB, Borda JT, Veazey RS, Myers L, Phillippi-Falkenstein KM, Bunnell BA, Ratterree MS, England JD. Serial electrophysiologic studies in rhesus monkeys with Krabbe disease. Muscle Nerve. 2005;32:185–90. doi: 10.1002/mus.20350. [DOI] [PubMed] [Google Scholar]
- Wenger DA. Murine, canine and non-human primate models of Krabbe disease. Mol Med Today. 2000;6:449–51. doi: 10.1016/s1357-4310(00)01800-1. [DOI] [PubMed] [Google Scholar]
- Wenger DA, Suzuki K, Suzuki Y, Suzuki K. Galactosylceramide Lipidosis: Globoid Cell Leukodystrophy (Krabbe Disease) In: Scriver CR, editor. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York: 2000. pp. 3669–3694. [Google Scholar]
- Wu YP, McMahon EJ, Matsuda J, Suzuki K, Matsushima GK. Expression of immune-related molecules is downregulated in twitcher mice following bone marrow transplantation. J Neuropathol Exp Neurol. 2001;60:1062–74. doi: 10.1093/jnen/60.11.1062. [DOI] [PubMed] [Google Scholar]
- Yagi T, McMahon EJ, Takikita S, Mohri I, Matsushima GK, Suzuki K. Fate of donor hematopoietic cells in demyelinating mutant mouse, twitcher, following transplantation of GFP+ bone marrow cells. Neurobiol Dis. 2004;16:98–109. doi: 10.1016/j.nbd.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Yamada H, Martin P, Suzuki K. Impairment of protein kinase C activity in twitcher Schwann cells in vitro. Brain Res. 1996;718:138–44. doi: 10.1016/0006-8993(96)00098-4. [DOI] [PubMed] [Google Scholar]
- Yamada H, Suzuki K. Responses to cyclic AMP is impaired in the twitcher Schwann cells in vitro. Brain Res. 1999;816:390–5. doi: 10.1016/s0006-8993(98)01142-1. [DOI] [PubMed] [Google Scholar]
- Yeager AM, Brennan S, Tiffany C, Moser HW, Santos GW. Prolonged survival and remyelination after hematopoietic cell transplantation in the twitcher mouse. Science. 1984;225:1052–4. doi: 10.1126/science.6382609. [DOI] [PubMed] [Google Scholar]
- Yeager AM, Shinohara M, Shinn C. Hematopoietic cell transplantation after administration of high-dose busulfan in murine globoid cell leukodystrophy (the twitcher mouse) Pediatr Res. 1991;29:302–5. doi: 10.1203/00006450-199103000-00016. [DOI] [PubMed] [Google Scholar]


