Abstract
Proteins containing a Tudor domain and domains homologous to staphylococcal nucleases are found in a number of eukaryotes. These “Tudor nucleases” have been found to be associated with the RNA-induced silencing complex (A. A. Caudy, R. F. Ketting, S. M. Hammond, A. M. Denli, A. M. Bathoorn, B. B. Tops, J. M. Silva, M. M. Myers, G. J. Hannon, and R. H. Plasterk, Nature 425:411-414, 2003). We have identified two Tudor nuclease gene homologs, TTN1 and TTN2, in the ciliate Tetrahymena thermophila, which has two distinct small-RNA pathways. Characterization of single and double KOs of TTN1 and TTN2 shows that neither of these genes is essential for growth or sexual reproduction. Progeny of TTN2 KOs and double knockouts occasionally show minor defects in the small-RNA-guided process of DNA deletion but appear to be normal in hairpin RNA-induced gene silencing, suggesting that Tudor nucleases play only a minor role in RNA interference in Tetrahymena. Previous studies of Tetrahymena have shown that inserted copies of the neo gene from Escherichia coli are often deleted from the developing macronucleus during sexual reproduction (Y. Liu, X. Song, M. A. Gorovsky, and K. M. Karrer, Eukaryot. Cell 4:421-431, 2005; M. C. Yao, P. Fuller, and X. Xi, Science 300:1581-1584, 2003). This transgene deletion phenomenon is hypothesized to be a form of genome defense. Analysis of the Tudor nuclease mutants revealed exceptionally high rates of deletion of the neo transgene at the TTN2 locus but no deletion at the TTN1 locus. When present in the same genome, however, the neo gene is deleted at high rates even at the TTN1 locus, further supporting a role for trans-acting RNA in this process. This deletion is not affected by the presence of the same sequence in the macronucleus, thus providing a counterargument for the role of the macronuclear genome in specifying all sequences for deletion.
Tudor nucleases.
Proteins characterized by the presence of four or five repeated domains with homology to staphylococcal nucleases and one Tudor domain are a class of conserved proteins found in a wide range of eukaryotes. The staphylococcal nuclease domains of Tudor nucleases have mutations in active-site residues such that nuclease activity is greatly reduced, although not entirely abolished (5, 6). Tudor domains are related to chromodomains and have been associated with DNA and RNA binding, methylated-histone binding (H3-K4 and H4-K20), and mediation of protein-protein interaction (5, 15, 16, 23, 31).
The functions of Tudor nucleases are not well understood, but some genetic and biochemical characterization in Drosophila melanogaster, Caenorhabditis elegans, mouse, and human cell lines, as well as Xenopus laevis oocytes, has provided clues to their biological functions. During biochemical characterization of the RNA-induced silencing complex (RISC) in Drosophila cell extracts, a 103-kDa protein containing the characteristic Tudor and staphylococcal nuclease domains, called Tudor-SN (TSN), was found to be associated with the Argonaute protein and other RNA interference (RNAi) machinery (6). It was originally hypothesized that this protein was the nuclease responsible for mRNA degradation by RISC, but the nuclease activity of RISC was subsequently found to be a function of the Argonaute protein, so the specific action of Tudor nuclease in the RISC complex is still unknown (20). RNAi against TSN in C. elegans led to defects in the function of the micro-RNA (miRNA) let-7 but had no effect on RNAi efficiency (6). Biochemical data show that TSN does have nuclease activity, despite mutations in nuclease active-site residues (6). A study with X. laevis oocyte extracts showed that Tudor nuclease binds to and promotes cleavage of hyperedited double-stranded RNAs which contain multiple inosine-uracil pairs (30). RNA editing and subsequent degradation by Tudor nuclease has also been linked to regulation of miRNA biogenesis (33). These studies suggest that Tudor nuclease may be part of an RNA editing pathway that could be another cellular mechanism for disposing of double-stranded RNAs.
Other functions in various organisms have been proposed, including roles in RNA transport, transcriptional coactivation, and splicing (5, 28, 32). Some of these functions are reasonably consistent with a role for Tudor nucleases in small RNA mediated processes, but the nature of these proteins' contributions is not clear.
RNAi and DNA deletion in Tetrahymena thermophila.
Tetrahymena has two small RNA pathways related to RNAi in other organisms (14). One pathway is responsible for double-stranded-RNA-induced gene silencing, which is mediated by 23- to 24-nucleotide (nt) small RNAs. The other pathway is involved in the genome rearrangement that occurs during sexual reproduction of this binucleate eukaryote (22, 24, 37). The macronucleus (MAC) of Tetrahymena contains around 50 copies each of approximately 200 different chromosomal fragments which are derived from the five chromosomes of the diploid micronucleus (MIC) by a process which eliminates 15% of the micronuclear genome (35). The eliminated sequences are known as deletion elements, or internal eliminated sequences, and are faithfully excised in every round of sexual reproduction. This process is thought to be guided by small RNAs, 28 to 30 nt in length, and requires several genes related to RNAi machinery genes, including the Argonaute gene homolog TWI1 and the Dicer gene homolog DCL1 (22, 24, 26, 34). However, the nuclease responsible for the actual cleavage of DNA during deletion has not been identified. The gene encoding Tudor nuclease, a nuclease associated with the RNAi machinery of the RISC complex, was an interesting candidate gene for study in T. thermophila. We have identified two genes in T. thermophila that have significant homology to Tudor nuclease genes found in other eukaryotes, which we refer to as TTN1 and TTN2 (for Tetrahymena Tudor nuclease 1 and 2). In this study, we began to characterize these genes to examine their possible roles in RNAi-mediated pathways.
Transgene deletion.
In addition to the normal DNA deletion that occurs during the sexual reproductive cycle, Tetrahymena has also been observed to delete a foreign sequence inserted into the micronuclear genome. Strains carrying an Escherichia coli neo gene in their MIC were found to produce progeny in which the neo gene had been deleted from the macronuclear chromosomes (21, 37). This process of transgene deletion was hypothesized to be a form of genome defense, but it is not fully understood how the cell determines what DNA is foreign and therefore to be eliminated. Deletion of the neo transgene has been previously observed at various genetic loci, and the frequency and extent of deletion have been found to vary depending on the locus and copy number (21, 37). We found an exceptionally high rate of neo deletion at the TTN2 locus but little or no deletion at the TTN1 locus. Such a high frequency of neo deletion from a single locus has not previously been described. When the TTN1 and TTN2 knockout (KO) strains were crossed to produce double KOs, this high deletion rate was also observed at the TTN1 locus. Analysis of transgene deletion at these two loci contributes new information to the study of this phenomenon.
MATERIALS AND METHODS
Northern blotting.
RNA samples were prepared using an RNeasy mini kit with Qiashredder (QIAGEN). Samples were combined 1:3 with NorthernMax formaldehyde load dye (Ambion) and electrophoresed on a 1.2% agarose formaldehyde MOPS gel. Gels were transferred to Hybond XL nylon membranes (Amersham Biosciences), cross-linked, and hybridized with probes overnight at 65°C in Church's hybridization buffer (1% [wt/vol] bovine serum albumin, 1 mM EDTA, 0.5 M phosphate buffer, 7% [wt/vol] sodium dodecyl sulfate [SDS]). Probes were made by random prime labeling of PCR products amplified from genomic DNA (see Table 1 for primer sequences) (29). After hybridization, blots were washed three times for 15 min each in SSC-0.1% SDS (SSC is 0.15 M NaCl, 0.015 M sodium citrate) and exposed to film.
TABLE 1.
Sequences of oligonucleotides used in Tudor nuclease experiments
| Primer use | Forward primer
|
Reverse primer
|
||
|---|---|---|---|---|
| Name | Sequence | Name | Sequence | |
| KO construction | ||||
| TTN1 5′ homology | ApaSN1for | CGGGGCCCGCCGCCTAAATTCAAAGAAA | XhoSN4rev | CCGCTCGAGTTTGGTAGAGTGGAGACCCTTC |
| TTN1 3′ homology | TetSN5 | GCACTTCAACTCATTGAAATCG | TetSN7-r | CCTTTTCAATAGCTTCATATTCGG |
| TTN2 5′ homology | Apa-TTN2-1 | GGGCCCTTTAAGACATTTCTTCGTCATCAA | Xho-TTN2-2r | CTCGAGTAGAGGCAGCCTGGTTTTCTATC |
| TTN2 3′ homology | TTN2-7 | CTTTTTGATCTAACAAATCCTTTAGC | TTN2-8R | AGACGGCCACAATAGAGCAT |
| Amplification of DNA for probes | ||||
| TTN1 probe | TetSN2 | AAAATAAATGGCTGCCCAAAA | TetSN4-r | TTTGGTAGAGTGGAGACCCTTC |
| TTN2 probe | Apa-TTN2-1 | GGGCCCTTTAAGACATTTCTTCGTCATCAA | Xho-TTN2-2r | CTCGAGTAGAGGCAGCCTGGTTTTCTATC |
| PCR analysis of KOs and neo deletion | ||||
| TTN1 KO vs WT | TetSN3 | ATGCTGAGTTCAATTTCGCC | TetSN5-r | CGATTTCAATGAGTTGAAGTGC |
| Mtt1rev | TCACAAATGATTAATGGGAGTCAAGG | |||
| TTN2 KO vs WT | TTN2-2 | GATAGAAAACCAGGCTGCCTCTA | TTN2-2.5rev | GCTAATTTTAATTCAATAGGAGGC |
| Mtt1rev | TCACAAATGATTAATGGGAGTCAAGG | |||
| TTN1 Δneo | MttDel3 | TTAGTGCACAATGTTTGAATGTT | TTN1NeoDel | GTGACACGTTTATAAGAATCTG |
| TTN2 Δneo | MttDel3 | TTAGTGCACAATGTTTGAATGTT | TTN2NeoDel | GTTTATAATATTTCAAAGCTAACAGCTTG |
Construction of TTN1 and TTN2 KO lines.
PCR primers (Table 1) were designed to amplify ∼1-kb regions of genomic DNA upstream and downstream of the region to be replaced by the neomycin resistance cassette. The upstream region was cloned into the ApaI and XhoI sites of the pMNBL(Neo3) vector, and the downstream region was cloned into the NotI and BamHI sites. Mating cultures of B2086II and CU428 cells (obtained from Cornell University through Peter Bruns) were transformed with KO constructs by particle bombardment as previously described (4). Transformants were selected in 120 μg/ml paromomycin. Micronuclear (germ line) transformants were identified by resistance to 6-methylpurine (6-Mp), as progeny of CU428 cells carry the 6-Mp resistance gene. Heterozygote germ line transformants were out-crossed to CU427 cells (obtained from Cornell University through Peter Bruns) to confirm heritability of the KO chromosome. For TTN1 germ line transformants, homozygotes were obtained by crossing two heterozygote strains and screening progeny by PCR to identify homozygotes. Two homozygote TTN1 KOs (lines 18 and 19) were obtained in this way. For TTN2 germ line KOs, homozygous strains were obtained using genomic exclusion crosses of heterozygous strains with star strains (13). Double KOs were constructed by crossing TTN1 homozygotes with TTN2 heterozygotes. Double-heterozygote KOs were identified by PCR screening, and these strains were used for genomic exclusion crosses to produce homozygous double KOs.
Progeny of TTN1, TTN2, and double KOs used in analysis of neo deletion were identified using cycloheximide or 6-Mp resistance markers or by testing potential progeny lines for remating.
Southern analysis of R-element DNA deletion.
For R-element analysis, DNA samples were digested with HindIII, electrophoresed in 1% agarose-Tris-borate-EDTA gels, and transferred to nylon membranes (29). Blots were hybridized overnight at 65°C with end-labeled probe (the 1.1-kb EcoRI/PstI fragment of the pDLCR5 plasmid [8]) in FBI buffer (1.5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA, pH 7.7], 10% polyethylene glycol, 7% SDS) (29). After hybridization, blots were washed three times for 20 min each at 65°C in 1× SSC and once for 20 min in 0.5× SSC and exposed to film.
RESULTS
T. thermophila has two Tudor nuclease genes.
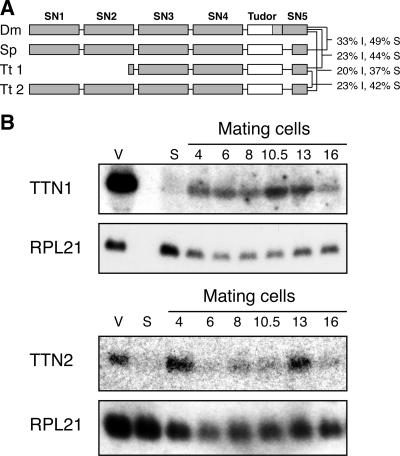
Two genes with homology to Tudor nuclease genes have been identified in the T. thermophila genome. TTN1 was initially identified using BLAST searches with Drosophila Tudor nuclease against published Tetrahymena cDNA sequences. This approach identified a partial cDNA, and more sequence was obtained by inverse PCR. Subsequently, the whole macronuclear genome of Tetrahymena was sequenced and the entire coding sequence of TTN1 was deduced from the genomic sequence (10). Upon further annotation of the genome sequence, a second gene with homology to Tudor nuclease genes was discovered, which we designated TTN2. Ttn1 is a truncated form of Tudor nuclease; translation of the cDNA predicts that it is 573 amino acids (aa) long and has only two complete SN domains (Fig. 1A). In contrast, Drosophila TSN (dTSN) is 926 aa long and contains four complete SN domains as well as the Tudor domain and another SN domain at the C terminus which partially overlaps the Tudor domain. The predicted Ttn2 protein is 800 aa long and aligns with the full length of dTSN and Tudor nucleases from other eukaryotes, having four SN domains, the Tudor domain, and another partial SN domain at the C terminus (Fig. 1A). When BLAST searches using these proteins were conducted, Ttn1 and dTSN showed 23% identity and 44% similarity, Ttn2 and dTSN showed 20% identity and 37% similarity, and Ttn1 and Ttn2 showed 23% identity and 42% similarity.
FIG. 1.
Tetrahymena Tudor nuclease proteins are similar to Tudor nucleases in other eukaryotes. (A) Schematic representation of Tudor nuclease proteins from D. melanogaster (Dm), Schizosaccharomyces pombe (Sp), and T. thermophila (Tt). Ttn1 is truncated compared to the other proteins, containing only two full nuclease domains. Brackets show percent identity and similarity between indicated proteins. (B) Expression of TTN1 and TTN2 in T. thermophila. Northern blots of RNA samples from vegetatively growing (v), starved (s), and mating cells at the indicated time points (in hours) after mixing were hybridized with probes to TTN1 and TTN2. A probe for RPL21 was used as a loading control. TTN1 shows the highest expression during vegetative growth. TTN2 shows peaks of expression during mating, at 4 and 13 h after mixing of cells.
Northern blots hybridized with a probe for TTN1 show that it is highly expressed in vegetatively growing cells, has very little expression in starved cells, and has weak expression throughout mating (Fig. 1B). TTN2 shows weak expression in vegetative growth, low expression in starved cells, and peaks of expression at 4 and 13 h postmixing during mating (Fig. 1B). Transcription of double-stranded RNAs from MIC-specific sequences begins at around 4 h postmixing during conjugation (9), and mRNA levels of other genes important for macronuclear development and DNA deletion, such as DCL1 and TWI1, peak between 2 h and 4 h as well (9, 22, 24, 26). On the other hand, small-RNA-mediated gene silencing occurs at all stages of the Tetrahymena life cycle (14). Therefore, the expression patterns of TTN1 and TTN2 are consistent with their possible participation in a RISC-like complex in Tetrahymena that could bind small RNAs.
TTN1 and TTN2 are not essential for growth or sexual reproduction.
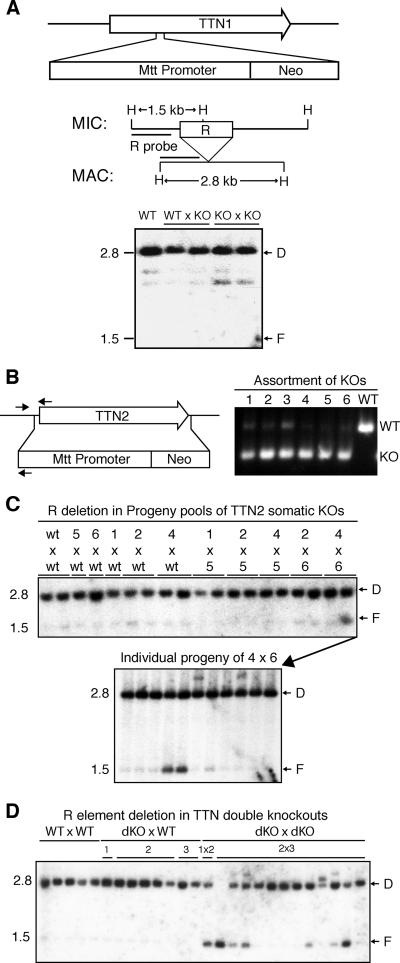
In order to determine if TTN1 was important for growth or conjugation in T. thermophila, the TTN1 gene was disrupted using transformation by homologous recombination. A neomycin resistance cassette (neo) was inserted between the first and second introns of the TTN1 gene, which resulted in KO strains with no detectable TTN1 transcript, as assayed by Northern blotting (data not shown). Both somatic (macronuclear) and germ line (micronuclear) transformants were obtained and analyzed. Growth of KO lines was normal, and matings of all KO lines showed normal progression and produced viable progeny (data not shown). Southern blotting analysis of DNA from TTN1 KO progeny showed that programmed DNA deletion occurred normally (Fig. 2A). These results led us to conclude that TTN1 is not required for growth or conjugation, including DNA deletion and MAC development.
FIG. 2.
Some progeny of TTN2 KO and double-KO cells show defects in R element deletion. (A) Diagram of the TTN1 locus showing insertion of the neo cassette in KO strains. Southern blotting analysis shows normal R deletion in progeny of TTN1 KO cells. A map of the R locus shows HindIII sites (H) and the region used as a probe in Southern analysis. Genomic DNA samples were made from pools of 8 to 12 individual progeny of the indicated matings. Positions of bands representing normal deletion (D; 2.8 kb) and failure of deletion (F; 1.5 kb) are indicated. (B) Diagram of TTN2 showing its complete replacement by the neo cassette in KO strains. Small arrows indicate positions of primers used in PCR analysis of KO chromosome assortment. Multiplex PCR on TTN2 somatic KO lines 1 to 6 shows that almost no wild-type (WT) chromosome remains in KO lines 1, 2, 4, 5, and 6 and only a small amount remains in line 3. (C) Southern blotting analysis of R-element deletion in progeny of somatic TTN2 KO lines was performed as for TTN1 KOs in panel A. One pool from the mating of KO lines 4 and 6 shows a significant amount of failure of deletion. Analysis of R deletion in genomic DNA from individual progeny lines of that pool shows that 2 of 11 individuals show a partial failure of R deletion. (D) Progeny of TTN1 and TTN2 double-KO cells also show defects in R-element deletion. Genomic DNA was made from individual progeny lines from matings between double-KO (dKO) lines 1 to 3 or wild-type (WT) lines. Southern analysis was performed as for panel C. Six of 13 progeny lines from double KOs show some failure of R deletion.
The TTN2 gene was disrupted by completely replacing the coding sequence with the neomycin resistance cassette (Fig. 2B). Somatic transformants with complete assortment to KO chromosomes were selected as previously described and analyzed by PCR (17) (Fig. 2B). Progeny from matings of selected lines were pooled in groups of 8 to 12 lines, and DNA deletion was analyzed by Southern blotting. For the most part, deletion occurred normally, with only two progeny lines from one mating showing significant failure in deletion of the R element (Fig. 2C). These results led us to conclude that expression of TTN2 in the parental MAC is not required for DNA deletion.
To determine if TTN1 and TTN2 have redundant functions in T. thermophila, double KOs were produced by crossing homozygous TTN1 KOs with heterozygous TTN2 KOs. Double heterozygote KOs were identified by PCR analysis and used to produce homozygous double KOs by using the methods of genomic exclusion crosses (13). These strains grew normally, indicating that these two genes are not essential for growth. Double-KO lines were mated with each other and with wild-type lines, and these matings were compared with matings of two wild-type lines. Matings of double KOs progressed normally and produced progeny comparable to those of the wild-type lines (data not shown). Individual progeny lines from KO and control matings were analyzed for DNA deletion. In one experiment, 6 of 13 progeny lines from double-KO matings showed a significant amount of failure of R deletion, compared to no failure in 13 lines from control matings (Fig. 2D). However, M deletion still occurred normally in these cells (data not shown). Subsequent matings of double KOs produced progeny that all showed normal deletion of R and M elements, indicating that the failure to delete the R element may be a phenotype of very low penetrance in these cells and may depend on some slight variation in conditions during mating. These results show that complete loss of both TTN1 and TTN2 does not significantly affect macronuclear development and DNA rearrangement in T. thermophila.
RNAi occurs normally in TTN KO cells.
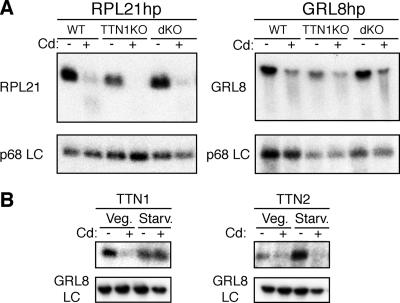
To determine if the Tudor nuclease genes are necessary for small-RNA-guided mRNA degradation, or RNAi, in Tetrahymena, germ line TTN1 and TTN1/TTN2 double KOs were transformed with RPL21 and GRL8 hairpin RNA expression constructs previously reported (14). Transformants were induced with Cd, and RNA samples were harvested and used for Northern analysis of target mRNAs. Target mRNA degradation occurred normally in both TTN1 KOs and TTN1/TTN2 double KOs (Fig. 3A). As in wild-type cells, expression of the hairpin RNA targeting the essential gene RPL21 in TTN1 or double KOs was lethal (data not shown).
FIG. 3.
TTN1 and TTN2 do not appear to function in RNAi in T. thermophila. (A) RNAi occurs normally in TTN1 and double KOs. Homozygous TTN1 knockouts (TTN1KO) or double KOs (dKO) were transformed with RPL21 and GRL8 hairpin constructs. Transformants were starved briefly and induced with Cd for 2 h, and then RNA was harvested. Northern blots were hybridized with probes for RPL21 and GRL8 to show mRNA degradation, and a p68 gene was used as a loading control. Both RPL21 and GRL8 hairpin transformants of WT, TTN1 KO, and double-KO lines show degradation of the target mRNA after induction with Cd. (B) Expression of TTN1 and TTN2 in SERH3 hairpin transformants. RNA samples from SERH3 hairpin-transformed cells in log growth (Veg.) or early starvation (Starv.), either untreated (−) or induced with Cd (+), were blotted and hybridized with probes for TTN1 or TTN2 to analyze expression; GRL8 was used as a loading control. Expression of hairpin RNA seems to cause decreases in levels of TTN1 in growing cells and of TTN2 in starved cells.
We have observed previously that when hairpin RNA expression is induced in Tetrahymena cells, the mRNA levels of genes involved in RNAi, such as Dicer and Argonaute gene homologs, are highly elevated (14). To determine if Tudor nuclease genes had similar responses, expression of TTN1 and TTN2 was examined in wild-type cells induced with Cd to express an RNA hairpin targeting the SERH3 gene. TTN1 levels actually decrease in growing cells expressing the hairpin but show little change in starved cells (Fig. 3B). TTN2 levels decrease in starved cells treated with Cd but show little change in growing cells. This response is the opposite of that seen for other genes thought to be involved in RNAi in Tetrahymena. These data support the hypothesis that the Tudor nuclease genes are not required for RNAi to function in T. thermophila and further argue that they are not likely to be involved in an ancillary role.
Deletion of the neo transgene in TTN2::neo KO cells is robust and independent of the presence of neo or TTN2 gene function in parental MAC.
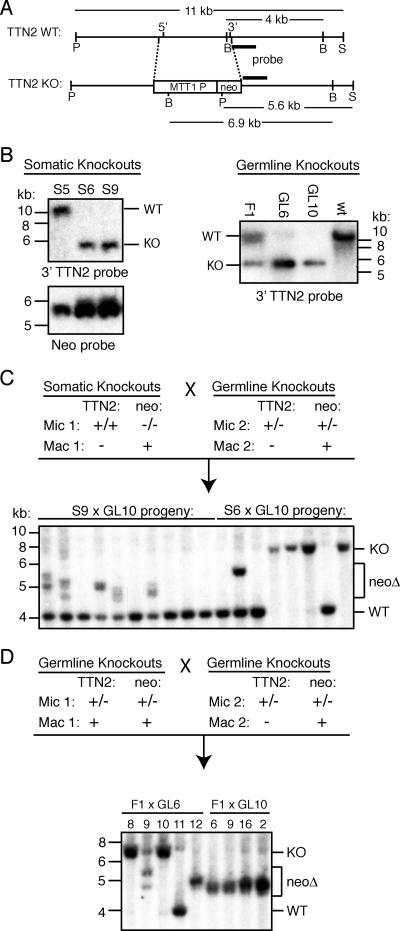
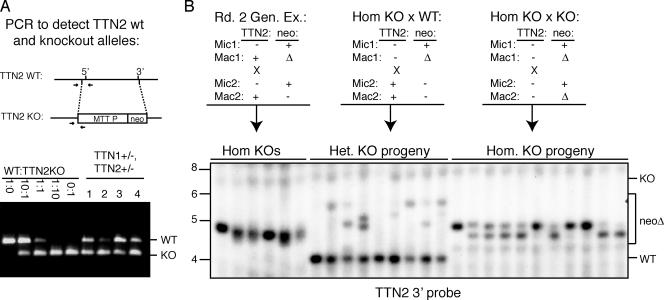
During analysis of the TTN2 KO strains in which the TTN2 coding sequence was replaced with the neo transgene under the control of the MTT1 promoter (Fig. 4A), we observed that the neo gene was frequently deleted in progeny of germ line transformants. This deletion was unusually efficient, unlike most previously reported deletions of the neo transgene at other gene loci in Tetrahymena. We chose to further investigate this deletion in order to address the nature of transgene deletion. It has been hypothesized that a transgene is recognized for deletion when it is present in the MIC but absent from the MAC or present at relatively low copy numbers, creating a situation similar to that observed with endogenous deletion elements (21, 37). However, this is apparently not the case here. Matings between somatic TTN2 KO lines, in which the MAC contained only the TTN2::neo chromosome, and heterozygous germ line KOs, which also had completely pure TTN2::neo MAC from assortment, frequently produced progeny in which the neo transgene was deleted (Fig. 4B and C). In 10 progeny of a mating between a somatic KO strain and a germ line heterozygote strain, all five of the progeny that inherited the KO chromosome showed evidence of neo deletion (Fig. 4C). In these matings, only one copy of the transgene is present in the parental MICs, and neo is present at high copy numbers in both parental MAC, yet neo deletion is robust. This indicates that deletion of a DNA sequence in the developing MAC does not depend on the absence of that sequence from the parental MAC. As would be expected, deletion of neo from the TTN2 locus is also robust in matings between two heterozygous germ line KOs (Fig. 4D), as well as matings between a homozygous germ line KO and a wild-type strain or between two homozygous germ line KO strains (Fig. 5B).
FIG. 4.
The neomycin resistance gene is frequently deleted from TTN2 KO cells, despite its presence in parental MAC. (A) Map of the TTN2 locus and KO gene structure. Letters beneath diagrams indicate restriction sites: P, PstI; B, BglII; S, StuI. Lengths of wild-type (WT) and KO restriction fragments detected in Southern hybridizations are indicated, and the location of the hybridization probe is shown by a black bar. (B) Southern hybridizations of PstI/StuI-digested genomic DNAs from somatic and heterozygous germ line KOs. Somatic KOs S6 and S9 and germ line KOs GL6 and GL10 show nearly complete assortment of the MAC to the KO chromosome. (C) Southern hybridization of BglII-digested genomic DNA from progeny of somatic KOs mated with germ line KOs. (Top) Diagram of the micronuclear and macronuclear genotypes of parental cell lines. (Bottom) Many progeny show BglII fragments smaller than the expected 6.9 kb, indicating deletion of the neomycin gene (neoΔ). (D) Southern blots of BglII-digested DNA from the progeny of heterozygous germ line KOs. Progeny chosen for Southern analysis were selected as homozygous KOs by PCR screening (except for no. 11, which was wild type). Many of the homozygous KOs show neo deletion bands, despite the fact that both parental MAC carried the neo gene.
FIG. 5.
neo deletion at the TTN2 locus is independent of TTN2 gene function. (A) PCR analysis of TTN1 and TTN2 double-heterozygote KOs show that MAC carry at least 50% wild-type TTN2 chromosomes. (Top) Location of primers used for PCR to detect KO and wild-type (WT) chromosomes. (Bottom) The first five lanes show results of control reactions with the indicated ratio of WT:TTN2 KO DNA. The last four lanes show reactions with DNA from double-heterozygote KOs, showing that all have wild-type TTN2. (B) Southern hybridization of BglII-digested DNA from homozygous KO progeny of round 2 genomic exclusion matings between double-heterozygote KOs in panel A and B*VI (lanes 1 to 6), heterozygous progeny of homozygous KOs mated with wild-type cells (lanes 7 to 15), and homozygous KO progeny of homozygous KO parents (lanes 16 to 26). As shown in the diagrams, both the round 2 genomic exclusion mating and the homozygous KO × WT mating show wild-type TTN2 gene function in at least one parental MAC. All matings shown produced progeny with neo deleted in the MAC, as indicated by the Δneo bands of 4.5 to 5.5 kb.
Because this high rate of deletion was observed in cells lacking a gene function (TTN2) that could possibly be involved in DNA deletion, it was important to determine if the TTN2::neo transgene deletion was correlated with the absence of TTN2 gene function in the parental MAC. To address this question, neo deletion was analyzed in TTN2 homozygous germ line KO strains and in progeny of TTN2 KO strains mated with wild-type strains. In both cases, the parental strains contained at least one MAC with a functional TTN2 gene.
TTN2 homozygous germ line KOs were produced by mating TTN1+/− TTN2+/− double heterozygote KOs with star strains in a process called genomic exclusion. In the first round of genomic exclusion, two cells types that have identical homozygous MICs, with one cell containing a wild-type MAC and the other the MAC of the original heterozygote parent, are created. In this case, the parental MAC of the heterozygotes contained at least 50% wild-type TTN2 chromosomes (Fig. 5A). In the second round of genomic exclusion, the two cells created in the first round (both MAC are TTN2+) are mated and produce homozygous MIC and MAC KO progeny. Homozygous TTN2 KOs were obtained from genomic exclusion matings by PCR screening of round 2 progeny to find strains that were homozygous KOs for TTN2 but wild-type for TTN1. Transgene deletion occurred in all homozygous KO progeny of round 2 genomic exclusion matings, indicating that the absence of TTN2 gene function is not required for robust neo deletion (Fig. 5). This is also the case in progeny of homozygous KOs mated with wild-type cells. These results indicate that TTN2::neo deletion is independent of TTN2 gene function.
The presence of TTN2::neo induces transgene deletion at the TTN1::neo locus.
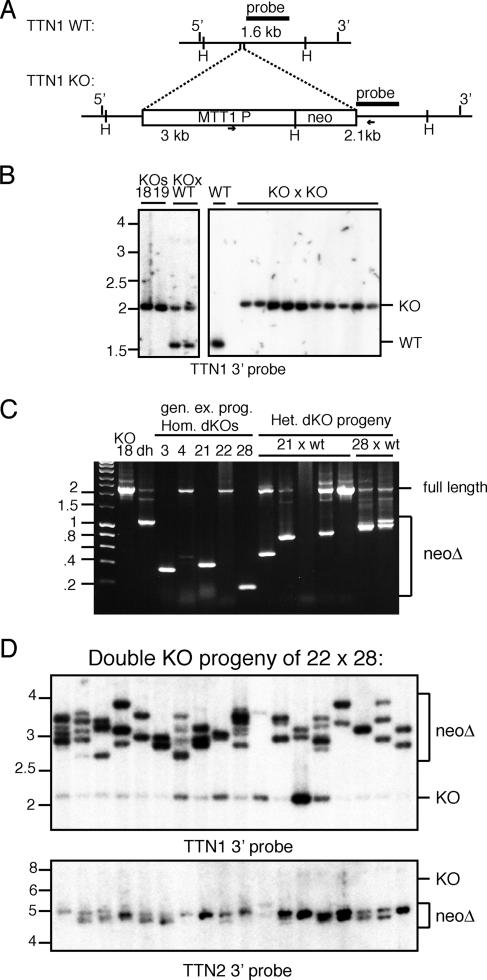
Unlike in TTN2 KOs, in TTN1 KOs, in which the same neo cassette was inserted to disrupt gene function (Fig. 6A), deletion of neo in progeny of homozygous germ line KOs was never observed (Fig. 6B). However, when the TTN1::neo and TTN2::neo strains were crossed to produce a double-KO heterozygote, robust deletion of neo at the TTN1 locus was observed (Fig. 6C). This TTN1::neo deletion was detected in the double heterozygote, in the homozygous double-KO progeny produced from it, and in the progeny of the double KOs mated with wild-type cells (Fig. 6C). When double KOs were mated with double KOs, 17 out of 18 progeny analyzed showed evidence of neo deletion at the TTN1 locus, and all showed complete neo deletion at the TTN2 locus (Fig. 6D). These results suggest that TTN2::neo somehow drives robust transgene deletion both at its own locus and at the TTN1 locus, which normally does not show neo deletion on its own.
FIG. 6.
Deletion of neo from the TTN1 locus is not detected in single KOs but is robust in double KOs. (A) Map of the TTN1 locus and KO gene structure. H, HindIII restriction sites. Lengths of wild-type (WT) and KO restriction fragments detected in Southern blots are indicated, and the location of the probe is shown by a black bar. Primers used for PCR analysis of neo deletion are indicated by small arrows. (B) Southern blots of HindIII-digested DNA of TTN1 homozygous KOs and their progeny show no evidence of neo deletion. (C) PCR analysis of neo deletion in TTN1 KO (lane 18), the TTN1+/− TTN2+/− double heterozygote (dh), the homozygous double KOs (lanes 3, 4, 21, 22, and 28), and progeny of homozygous KOs mated with wild-type cells shows that there are neo deletion events at the TTN1 locus in many progeny of matings where neo is present at the TTN2 locus. (D) Southern blots of DNAs of progeny of double-KO cells. HindIII-digested DNAs were probed with the TTN1 3′ flank, and BglII-digested DNAs were probed with the TTN2 3′ flank. Deletion of neo from the TTN1 locus results in the loss of a HindIII site, resulting in larger DNA fragments (between 2.5 and 4 kb) (neoΔ). Blots show that nearly all double homozygous KO progeny show substantial deletion of neo from the TTN1 locus and complete deletion of neo from the TTN2 locus.
TTN1::neo and TTN2::neo transgene deletions have similar lengths and borders.
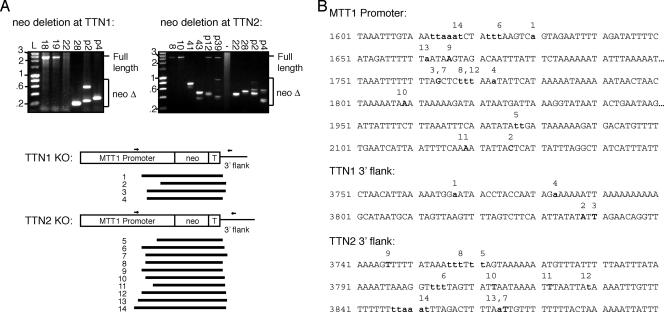
To examine the borders of deletion at the TTN1::neo and TTN2::neo loci, PCR primers were designed to amplify across the region of transgene deletion (Fig. 7A). These PCR products were then cloned and sequenced to determine the borders of transgene deletion, which are listed in Table 2. Most of the deletions sequenced were 1.8 to 2.2 kb in length, and they included the neo gene, the 3′ BTU2 termination sequence, 50 to 150 bases of 3′ TTN1- or TTN2-flanking sequence, and up to nearly 1 kb of the MTT promoter (Fig. 7). It must be noted, however, that deletions larger than 2.4 kb could not be amplified by the primers used in this experiment. The sizes of the deletions and the deletion of a large part, but not all, of the MTT promoter are interesting. Previously reported deletions are generally about 1 kb in length and sometimes include shorter promoter regions (such as the 330-bp H4-I promoter) and terminator regions (21, 37). The deletion of such a large part of the MTT promoter has not been previously described in cases of neo deletion from KO cassettes that employ this promoter (21). It is interesting to speculate whether this “extra” deletion is due to the robustness of deletion occurring in these strains or is somehow a function of the chromatin structure that occurs at these particular loci. The borders of deletion were more variable on the 5′ end, in the MTT promoter region, but often deletions in two different strains shared a border on one end or the other, indicating that there may be “hot spots” that are preferentially chosen as borders (Fig. 7B). Deletion borders often contain ambiguous bases (bases that are shared by the 5′ and 3′ ends), indicating that microhomologies may contribute to border choice, as has been previously observed in other deletion events (14) (Fig. 7B).
FIG. 7.
Analysis of deletion borders in TTN2 KO and double-KO strains. (A) DNA from TTN2 KO and double-KO strains was used for PCR amplification across the neo cassette by primers (small arrows). PCR products smaller than the expected full length were indicative of neo deletion and were cloned for sequencing. The diagram shows the extent of DNA deletion in analyzed strains. Strains and deletions correspond to those listed in Table 2. (B) Sequences of DNA regions where deletion borders occur in the MTT promoter, TTN1 3′ flank, and TTN2 3′ flank. Deletion borders are in boldface. Numbers above sequences correspond to deletion border sequences in panel A and Table 2. Deletion borders that contain ambiguous bases are designated with lowercase letters in both the MTT promoter and 3′ flank sequences, as it is not possible to distinguish the origin of the base.
TABLE 2.
Sequences of deletion junctions in TTN1 and TTN2 KO strains
| Deletion site and strain | Sequence of deletion junctiona |
|---|---|
| TTN1 | |
| TTNdKO 28-3 | AATCTATTTAAGTCA/ATAACCTACCAATAG |
| TTNdKOp 2C | TTTCAAAATATTAC/ATTAGAACAGGTTTAT |
| TTNdKOp 2C | TTAAATTTTTTTTAG/TAGAACAGGTTTATA |
| TTNdKOp 4C | TTTTTTTAGCTCTTTAAaAAAAATTAAAAA |
| TTN2 | |
| TTN2KO 41 | AATTTCAAATATAttAGTAAAAAAATGTTT |
| TTN2KO 43 | AAATTAAATCTAtttTAAAGGTTTTTAGTT |
| TTN2KOp 12 | TTAAATTTTTTTTAG/TTGTTTTTTTTACTA |
| TTN2KOp 39 | AAATTTTTTTTAGCTCtttTTTAGTAAAAA |
| TTNdKO 22-1 | TAGATTTTTTAATAA/TTTTTATAAATTTTT |
| TTNdKO 28-3 | ACTAACTAAAAATAA/TAATAAAATTTAATT |
| TTNdKOp 2C | TCATTAATTTTCAAA/TTAATTATAAAATTT |
| TTNdKOp 2C | TTTTTAGCTCTTtAAAATTTGTTTTTTTTT |
| TTNdKOp 4C | TTTCATAGATTTTTTaTTGTTTTTTTTACT |
| TTNdKOp 4C | TTGATAAATTTGTAAAttaaatTTAGACTTTTAAT |
Lowercase letters indicate bases with ambiguous origins as described in the legend to Fig. 7B.
DISCUSSION
In this study, we examined the effects of loss of function of two Tudor nuclease genes in T. thermophila. Although RNAi has been performed against TSN in C. elegans, no KO of Tudor nuclease genes has been previously reported. TTN1 and TTN2 KO cell lines are viable and did not display severe defects in macronuclear development, programmed DNA deletion, or RNA-guided gene silencing. These results show that Tudor nuclease genes are not essential in Tetrahymena. The phenotypes of the KO cells are somewhat inconsistent with previous work that implicated Tudor nuclease as a RISC complex protein necessary for proper miRNA function (6). However, in Tetrahymena, a group of endogenous 23- to 24-nt RNAs that correspond to the antisense strand of several predicted gene clusters have been identified and sequenced (19). These RNAs are produced by the coupled action of an RNA-dependent RNA polymerase (Rdr1) and a Dicer RNase (Dcr2) (18), but the function of these RNAs is currently unknown. Therefore, it is possible that TTNs may be involved in this small-RNA pathway, which has yet to be fully characterized. Biochemical work is needed to determine if either TTN1 or TTN2 associates with other proteins involved in small RNA pathways, such as the Argonaute homologs (TWIs) or Dicers. Repeated BLAST searches of the Tetrahymena genome have not revealed any other candidate Tudor nucleases; therefore, it is unlikely that another Tudor nuclease could be required for RNAi or DNA deletion.
Tetrahymena cells lacking both TTN1 and TTN2 function did show some minor defects in deletion of one specific element, the R deletion element. In past studies, the R element has been shown to be more sensitive to perturbation, and thus more likely to show failure of deletion, than other DNA deletion elements (8). Therefore, it may be a more sensitive indicator of slight defects in the DNA deletion process than other DNA deletion elements. Defects in R deletion in Tudor nuclease KO lines may therefore point to a role for these genes that is not essential for pathway function but rather increases fidelity or efficiency of the DNA deletion process. No other genes in Tetrahymena have been shown to be required for deletion of specific elements or classes of elements, although this phenomenon has been reported in Paramecium (27). Deletion elements in Tetrahymena vary in size, border fidelity, and copy number and can be grouped into several different classes based on these attributes. It is likely that deletion of different classes of elements will require different protein factors. The R element is similar in size and border fidelity to the nearby M element but does not share significant sequence homology (1-3). It seems probable that they would have similar molecular requirements for proper deletion, but it is technically possible that one would require additional factors. This could explain the difference in deletion fidelity between the R and M elements in cells lacking Tudor nucleases.
Additional experiments were performed in an attempt to reveal other phenotypes associated with loss of Tudor nucleases in Tetrahymena cells. Experiments described in this study indicate that TTNs are not required for RNAi in Tetrahymena. The TTN1 KO, TTN2 KO, and double-KO lines were also subjected to a variety of extreme growth conditions, including high and low temperatures, media of various pHs and osmotic strengths, and chemical stresses such as caffeine. However, no difference in tolerance of these conditions was seen between wild-type and KO lines (data not shown).
It is possible that Tudor nucleases in Tetrahymena could be involved in degradation of hyperedited RNAs, as has been reported for an X. laevis oocyte system (30). Such a function could be linked to the endogenous 23- to 24-nt small-RNA pathway, as such a linkage has been reported for RNA editing and micro-RNA biogenesis in a mammalian system (33). RNA editing processes have not been previously investigated in Tetrahymena, so this could be a new avenue for future study.
The variety of functions that have been proposed for Tudor nucleases in different organisms (RISC complex member, transcriptional coactivator, edited dsRNA nuclease) suggest that these proteins may have evolved different functions in different organisms. Alternatively, Tudor nucleases may act as general RNA binding adaptor proteins that function in many different processes in the cell. Such a function would be consistent with all of the currently recorded interactions of Tudor nucleases. In Tetrahymena, such functions may be redundant with those of other genes, since Tudor nucleases are not essential to growth in normal laboratory conditions. However, free-living Tetrahymena cells encounter many conditions that are difficult to reproduce in a laboratory setting. Tudor nucleases may be used in response to environmental stresses. It is hoped that future experiments will provide a clearer understanding of the biological function of this family of conserved genes.
Investigation of transgene deletion at the TTN1 and TTN2 loci has led to new information concerning this phenomenon. A proposed function of DNA deletion (including transgene deletion) in Tetrahymena is as a mechanism of genome defense (37). Tetrahymena cells are able to “lose” detrimental DNA sequences from their somatic genome by the process of assortment, which occurs in the polyploid, amitotically dividing MAC. However, any unwanted or foreign sequences that invade the MIC will be transmitted to the MAC of future progeny. DNA deletion can serve as a means to prevent this transmission, as the undesirable sequences are eliminated during development of the progeny's new MAC during sexual reproduction. It is not fully understood how the cell knows what DNA to delete and what to keep. Based on evidence from studies of endogenous elements, a model has been proposed called the “scan RNA” model for DNA elimination (21, 24, 25). This model proposes that all sequences in the MIC genome are bidirectionally transcribed early in conjugation and the transcripts are processed into small RNAs, which are scanned against the old MAC, eliminating RNAs that match sequences found in the old MAC. The remaining RNAs then go on to the developing new MAC and target DNA sequences for elimination. In this way, sequences that are in the MIC but not in the old MAC are eliminated from the developing new MAC. This model was based partially on earlier evidence that the presence of a normally deleted sequence in the MAC can prevent its deletion in the next round of sexual reproduction (8). In this study, we show that robust transgene deletion of neo can occur regardless of the presence or absence of neo in the parental MAC. Thus, comparison between the MAC and MIC sequences through scanning RNA cannot explain this deletion. Clearly then, the cell has a different mechanism to determine what DNA is “self” and what is “foreign.” Although repeated efforts to find commonalities in sequences of deletion elements have failed, there may be specific properties of eliminated sequences at a more structural level that the cell identifies as foreign.
In addition, the cell's ability to “see” a transgene and designate it as foreign varies according to the location of the transgene in the genome. Previous studies have shown that transgenes at some loci are deleted fairly frequently, while transgenes at other loci are rarely deleted (21, 37). The two loci examined in this study, TTN1 and TTN2, fall at opposite ends of this spectrum. It has been reported that the deletion of neo at “resistant” loci can be increased by increasing the total copy number of neo in the parental MICs, but in one case three copies per haploid genome in each mating partner were needed to cause deletion (21). The present study shows that deletion of neo from a resistant locus can be driven by just one additional copy of neo at a locus that promotes efficient deletion, as is the case with TTN2::neo driving deletion of TTN1::neo. This observation provides strong support for the argument that neo deletion, like the deletion of endogenous deletion elements, is driven by a trans-acting substance (such as the double-stranded RNA) produced from this locus (21). Thus, the difference in deletion efficiency between TTN1 and TTN2 likely reflects the amounts of RNA they are able to produce and not the physical constraints of the locus on the machinery of deletion.
In T. thermophila, the borders of DNA elimination of some deletion elements are very precise, while others are more variable, and how these borders are determined is not well understood (36). The precise borders of some elements are determined by flanking regulatory sequences, but the regulatory sequences differ from element to element (7, 11, 12). In the deletion of the transgene neo, border selection seems somewhat arbitrary. Deletion borders often lie near, but not precisely on, the boundaries between the transgene and the flanking Tetrahymena DNA. However, “misplaced” Tetrahymena DNA, such as the promoter and terminator sequences used to create the neo expression cassette, is sometimes deleted and sometimes left behind (21, 37). In the case of the TTN1::neo and TTN2::neo deletions, the terminator sequence and about half of the MTT promoter sequence are eliminated at both loci. It is possible that there is an internal sequence in the MTT promoter that acts like a border determination sequence, causing the deletion to stop in the middle. However, this boundary was not reported for deletions occurring at another locus containing the same KO cassette (21). The similarity in deletion borders in the two loci, and the appearance that TTN2::neo deletion drives TTN1::neo deletion in the double KO, suggests that some mechanism, at either the RNA or DNA level, links the transgene deletion at these two loci. Previous results showed that transcripts produced from deletion elements have heterogeneous ends that do not correspond closely with deletion borders (9). In addition, DNA deletion induced by injection of homologous RNAs or expression of hairpin RNAs often spans a larger region than that corresponding to the targeting RNAs (14, 37). Thus, it seems unlikely that RNAs alone are the major deciding factor in border choice. This leaves the possibility that the DNA sequences at the two loci somehow interact, perhaps via the shared sequences in the neo KO cassette.
This study provides new insight into the process of transgene elimination in T. thermophila. Clearly, there are still many details that are not fully understood. The TTN1 and TTN2 loci, which show dramatically different levels of neo deletion, will be useful in future studies of transgene deletion that address issues of transcription and small-RNA production, identification of foreign sequence versus self sequence, and border determination.
Acknowledgments
We express our thanks to Teresa Bleakly for help with creating KO constructs and to Jonathan Myers, Tessa Robinson, Jerill Thorpe, and Aida Flor de la Cruz for help in analysis of KO phenotypes. We are grateful to all members of the Yao lab for helpful discussions and encouragement.
This work was supported by National Institutes of Health grant GM26210 and Academia Sinica.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Austerberry, C. F., C. D. Allis, and M. C. Yao. 1984. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc. Natl. Acad. Sci. USA 81:7383-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austerberry, C. F., and M. C. Yao. 1987. Nucleotide sequence structure and consistency of a developmentally regulated DNA deletion in Tetrahymena thermophila. Mol. Cell. Biol. 7:435-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austerberry, C. F., and M. C. Yao. 1988. Sequence structures of two developmentally regulated, alternative DNA deletion junctions in Tetrahymena thermophila. Mol. Cell. Biol. 8:3947-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruns, P. J., and D. Cassidy-Hanley. 2000. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 62:501-512. [DOI] [PubMed] [Google Scholar]
- 5.Callebaut, I., and J. P. Mornon. 1997. The human EBNA-2 coactivator p100: multidomain organization and relationship to the staphylococcal nuclease fold and to the Tudor protein involved in Drosophila melanogaster development. Biochem. J. 321:125-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caudy, A. A., R. F. Ketting, S. M. Hammond, A. M. Denli, A. M. Bathoorn, B. B. Tops, J. M. Silva, M. M. Myers, G. J. Hannon, and R. H. Plasterk. 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425:411-414. [DOI] [PubMed] [Google Scholar]
- 7.Chalker, D. L., A. La Terza, A. Wilson, C. D. Kroenke, and M. C. Yao. 1999. Flanking regulatory sequences of the Tetrahymena R deletion element determine the boundaries of DNA rearrangement. Mol. Cell. Biol. 19:5631-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalker, D. L., and M. C. Yao. 1996. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol. Cell. Biol. 16:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalker, D. L., and M. C. Yao. 2001. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 15:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisen, J. A., R. S. Coyne, M. Wu, D. Wu, M. Thiagarajan, J. R. Wortman, J. H. Badger, Q. Ren, P. Amedeo, K. M. Jones, L. J. Tallon, A. L. Delcher, S. L. Salzberg, J. C. Silva, B. J. Haas, W. H. Majoros, M. Farzad, J. M. Carlton, R. K. Smith, Jr., J. Garg, R. E. Pearlman, K. M. Karrer, L. Sun, G. Manning, N. C. Elde, A. P. Turkewitz, D. J. Asai, D. E. Wilkes, Y. Wang, H. Cai, K. Collins, B. A. Stewart, S. R. Lee, K. Wilamowska, Z. Weinberg, W. L. Ruzzo, D. Wloga, J. Gaertig, J. Frankel, C. C. Tsao, M. A. Gorovsky, P. J. Keeling, R. F. Waller, N. J. Patron, J. M. Cherry, N. A. Stover, C. J. Krieger, C. del Toro, H. F. Ryder, S. C. Williamson, R. A. Barbeau, E. P. Hamilton, and E. Orias. 2006. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4:e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godiska, R., C. James, and M. C. Yao. 1993. A distant 10-bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes Dev. 7:2357-2365. [DOI] [PubMed] [Google Scholar]
- 12.Godiska, R., and M. C. Yao. 1990. A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell 61:1237-1246. [DOI] [PubMed] [Google Scholar]
- 13.Hai, B., J. Gaertig, and M. A. Gorovsky. 2000. Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 62:513-531. [DOI] [PubMed] [Google Scholar]
- 14.Howard-Till, R. A., and M. C. Yao. 2006. Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Mol. Cell. Biol. 26:8731-8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, Y., J. Fang, M. T. Bedford, Y. Zhang, and R. M. Xu. 2006. Recognition of histone H3 lysine-4 methylation by the double Tudor domain of JMJD2A. Science 312:748-751. [DOI] [PubMed] [Google Scholar]
- 16.Iwabuchi, K., B. P. Basu, B. Kysela, T. Kurihara, M. Shibata, D. Guan, Y. Cao, T. Hamada, K. Imamura, P. A. Jeggo, T. Date, and A. J. Doherty. 2003. Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J. Biol. Chem. 278:36487-36495. [DOI] [PubMed] [Google Scholar]
- 17.Karrer, K. M. 2000. Tetrahymena genetics: two nuclei are better than one. Methods Cell Biol. 62:127-186. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. R., and K. Collins. 2007. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat. Struct. Mol. Biol. 14:604-610. [DOI] [PubMed] [Google Scholar]
- 19.Lee, S. R., and K. Collins. 2006. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev. 20:28-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, J., M. A. Carmell, F. V. Rivas, C. G. Marsden, J. M. Thomson, J. J. Song, S. M. Hammond, L. Joshua-Tor, and G. J. Hannon. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437-1441. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Y., X. Song, M. A. Gorovsky, and K. M. Karrer. 2005. Elimination of foreign DNA during somatic differentiation in Tetrahymena thermophila shows position effect and is dosage dependent. Eukaryot. Cell 4:421-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malone, C. D., A. M. Anderson, J. A. Motl, C. H. Rexer, and D. L. Chalker. 2005. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol. Cell. Biol. 25:9151-9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurer-Stroh, S., N. J. Dickens, L. Hughes-Davies, T. Kouzarides, F. Eisenhaber, and C. P. Ponting. 2003. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28:69-74. [DOI] [PubMed] [Google Scholar]
- 24.Mochizuki, K., N. A. Fine, T. Fujisawa, and M. A. Gorovsky. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110:689-699. [DOI] [PubMed] [Google Scholar]
- 25.Mochizuki, K., and M. A. Gorovsky. 2004. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 18:2068-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mochizuki, K., and M. A. Gorovsky. 2005. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 19:77-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowacki, M., W. Zagorski-Ostoja, and E. Meyer. 2005. Nowa1p and Nowa2p: novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia. Curr. Biol. 15:1616-1628. [DOI] [PubMed] [Google Scholar]
- 28.Paukku, K., J. Yang, and O. Silvennoinen. 2003. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol. Endocrinol. 17:1805-1814. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning, a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Scadden, A. D. 2005. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 12:489-496. [DOI] [PubMed] [Google Scholar]
- 31.Sprangers, R., M. R. Groves, I. Sinning, and M. Sattler. 2003. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J. Mol. Biol. 327:507-520. [DOI] [PubMed] [Google Scholar]
- 32.Yang, J., T. Valineva, J. Hong, T. Bu, Z. Yao, O. N. Jensen, M. J. Frilander, and O. Silvennoinen. 2007. Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Res. 35:4486-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, W., T. P. Chendrimada, Q. Wang, M. Higuchi, P. H. Seeburg, R. Shiekhattar, and K. Nishikura. 2006. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 13:13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao, M. C., and J. L. Chao. 2005. RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu. Rev. Genet. 39:537-559. [DOI] [PubMed] [Google Scholar]
- 35.Yao, M. C., J. Choi, S. Yokoyama, C. F. Austerberry, and C. H. Yao. 1984. DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell 36:433-440. [DOI] [PubMed] [Google Scholar]
- 36.Yao, M. C., S. Duharcourt, and D. L. Chalker. 2002. Genome-wide rearrangements of DNA in ciliates, p. 730-758. In N. L. Craig, R. Craigie, M. Gillert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 37.Yao, M. C., P. Fuller, and X. Xi. 2003. Programmed DNA deletion as an RNA-guided system of genome defense. Science 300:1581-1584. [DOI] [PubMed] [Google Scholar]